Investigation of novel SARS-CoV-2 variant
Variant of Concern 202012/01
Technical briefing 4
This briefing provides an update on the briefing of 8 January 2021
Nomenclature of variants in the UK
SARS-CoV-2 variants if considered to have concerning epidemiological, immunological or
pathogenic properties are raised for formal investigation. At this point they are designated
Variant Under Investigation (VUI) with a year, month, and number. Following risk assessment
with the relevant expert committee, they may be designated Variant of Concern (VOC). This
variant was designated VUI 202012/01 on detection and on review re-designated as VOC
202012/01 on 18 December 2020.
Virology
The workflow in process by PHE and partner laboratories is shown in Figure 1.
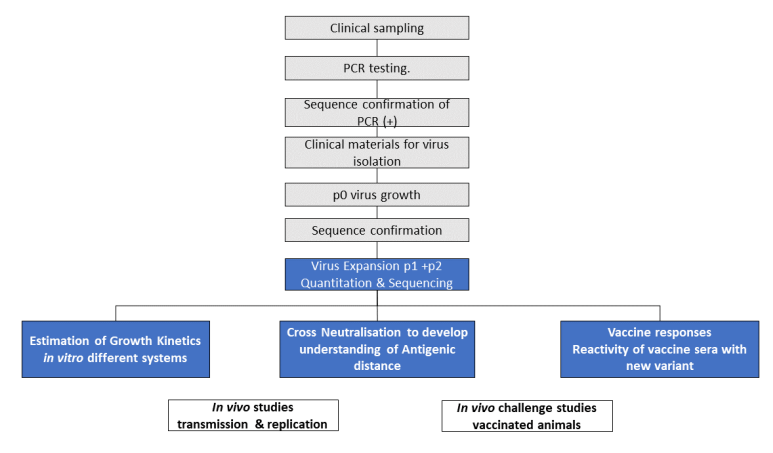
NB: Activities on grey background (black text) are completed and on blue background (white text) are underway.
Text alternative to Figure 1 workflow:
1. Clinical sampling
2. PCR testing
3. Sequence confirmation of PCR (+)
4. Clinical materials for virus isolation
5. p0 virus growth
6. Sequence confirmation
7. Virus Expansion p1 +p2 Quantitation & Sequencing
i. Estimation of Growth Kinetics in vitro different systems
ii. Cross Neutralisation to develop understanding of Antigenic distance
iii. Vaccine responses Reactivity of vaccine sera with new variant
8. In vivo work
i. In vivo studies transmission & replication
ii. In vivo challenge studies vaccinated animals
NB: Activities 1-6 are completed and activity 7 is underway.
1. B1.1.7 virus demonstrates differences in growth characteristics when compared to a selection
of viruses from UK cases using in vitro systems. B1.1.7 grows well in human airway epithelium and
less well in Vero cell lines, when compared to all other isolates tested so far. (Moderate to High
confidence)
2. Neutralisation studies have commenced in 3 different laboratories using a small number of
different B1.1.7 isolates and comparator viruses and will shortly begin in a fourth.
Findings to date are:
• consistent evidence of cross-neutralising activity in convalescent sera. Sera from individuals
who have been infected with non-B1.1.7 lineages show neutralising activity against B1.1.7
virus, and the converse is also true. (moderate to high confidence)
• across experiments from several laboratories there is evidence suggesting antigenic
distance between B1.1.7 and the other viruses tested, however, further investigations are
needed to better quantify the effect size and determine the significance
• neutralisation studies using post-vaccination sera are still underway
S gene target failure/lineage correlation
Only a small fraction of all new cases of VOC 202012/01 are identified by whole-genome
sequencing, and this data typically lags test date by approximately 2 weeks, therefore a proxy S
gene target failure (SGTF) is used to monitor the VOC.
We previously observed that one of the S gene mutations in the VOC, which deletes amino
acids 69 and 70 (Δ69-70), causes a reproducible S gene target failure (SGTF) in the
Thermopath TaqPath assay used in 3 UK lighthouse laboratories (see Technical Briefing 1).
This coincidental occurrence provides a good proxy for monitoring trends in VOC 202012/01.
SGTF correlates almost perfectly with presence of Δ69-70. Considering 23,428 tested samples
where we know both the sequence and the SGTF status, 99.6% of Δ69-70 sequences (6641 of
6669) are SGTF, compared to 0.05% of sequences without the deletion (9 of 16759).
Because Δ69-70 has arisen multiple times, and SGTF is a proxy for any lineage with that
mutation, the utility of SGTF as a proxy for VOC 202012/01 varies over time and region. Table 1
shows, for all pillar 2 sequences, the weekly proportion of Δ69-70 sequences that were
confirmed to be VOC 202012/01. Table 2 shows the proportion of Δ69-70 that is the VOC
202012/01 in England since December 1, broken down by region. It is, as expected, highest in
the areas where the VOC was first observed, but it has been a substantial majority in all areas
of England during the month of December. The numbers in these tables are based on
sequenced samples, some of which may have come from the same individual (this effect is
likely to be small).
| Week beginning | Per cent VOC of all Δ69-70 | Number of pillar 2 Δ69-70 sequences |
| 2020-10-12 | 3% | 116 |
| 2020-10-19 | 15% | 219 |
| 2020-10-26 | 29% | 156 |
| 2020-11-02 | 64% | 399 |
| 2020-11-09 | 81% | 712 |
| 2020-11-16 | 89% | 770 |
| 2020-11-23 | 93% | 387 |
| 2020-11-30 | 95% | 442 |
| 2020-12-07 | 98% | 2698 |
| 2020-12-14 | 99% | 3988 |
| 2020-12-21 | 99% | 2203 |
| 2020-12-28 | 100% | 238 |
| Region | Percent VOC 202012/01 of all Δ69-70 | Number of pillar 2 Δ69-70 1 December to 3 January |
| East Midlands | 91% | 162 |
| East of England | 99% | 1787 |
| London | 99% | 3614 |
| North East | 96% | 264 |
| North West | 98% | 537 |
| South East | 99% | 2300 |
| South West | 100% | 174 |
| West Midlands | 98% | 428 |
| Yorkshireand the Humber | 94% | 191 |
Analysis of SGTF data between 1 October 2020 and 31 December 2020 of matched SGTF data to
COGUK sequence data including 24,727 sequences from Pillar 2 specimens tested by the TaqPath
laboratories.
Figure 2 shows the weekly percentage of VOC202012/01 and STGF combinations and shows
increasing proportion of VOC202012/01 with SGTF (data for England only where PHE region is
assigned). Unknown category in Figure 2 comprises COGUK identifiers not within the EDGE
database at current time or sequences wherein sequence quality is insufficient to assign variant.
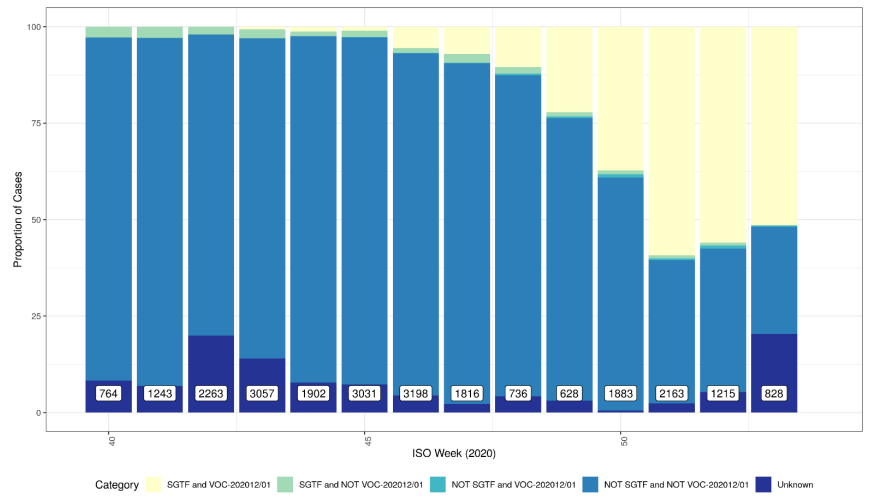
Data for England (not UK), no deduplicated. Category combinations include SGTF and VOC 202012/01 (Sample with S gene
target failure classified as a VOC-202012/01); SGTF and NOT VOC 202012/01 (Sample with S gene target failure NOT classified
as a VOC-202012/01); NOT SGTF and VOC 202012/01 (Sample did not have S gene target failure, but is classified as VOC202012/01);
NOT SGTF and NOT VOC 202012/01 (Sample did not have S gene target failure, and is NOT classified as VOC202012/01);
Unknown (Unknown VOC 202012/01 status).
Epidemiology of S gene target failure
The proportion of England specimens tested in the lighthouse laboratories using the assay
which produces the S gene target failure is substantial and has been relatively constant over time
(Figure 3). This however varies by geography, with lower coverage between 1 September 2020
and 11 January 2021 in local authorities in the East and South West of England.
The proportion of cases tested by this assay who have isolates with SGTF has continued to rise in
December (Figure 4 and 5) in all regions (Figure 6). In the most recent seven-day period (5 to 11
January 2021), 76.6% of 123,652 Pillar 2 cases detected in TaqPath laboratories had isolates with
SGTF, compared to 71.7% in the prior seven-day period and 27.7% in the seven-day period
starting 1 December 2020.
The spatial distribution of SGTF cases shows a relatively higher burden in the south of England
but with clear evidence of spread across other areas, particularly in the North West (Figure 4).
Cases by age and sex are displayed (Figure 7 and 8). Cases that had isolates with SGTF
display a similar age-sex profile to other Pillar 2 cases tested by the TaqPath laboratories since
1 September 2020. Proportions of cases with SGTF were also similar across those aged under
25, between 25 and 59, and over 60: 76.2%, 77.1%, and 74.5% respectively among those
diagnosed by TaqPath laboratories in the most recent seven-day period.
Data on coverage of TaqPath Lab testing and numbers/proportions of cases with SGTF are
shared daily with Local Authorities (Sunday-Friday) on the COVID-19 PHE Local Authorities
Report Store (Sharepoint).
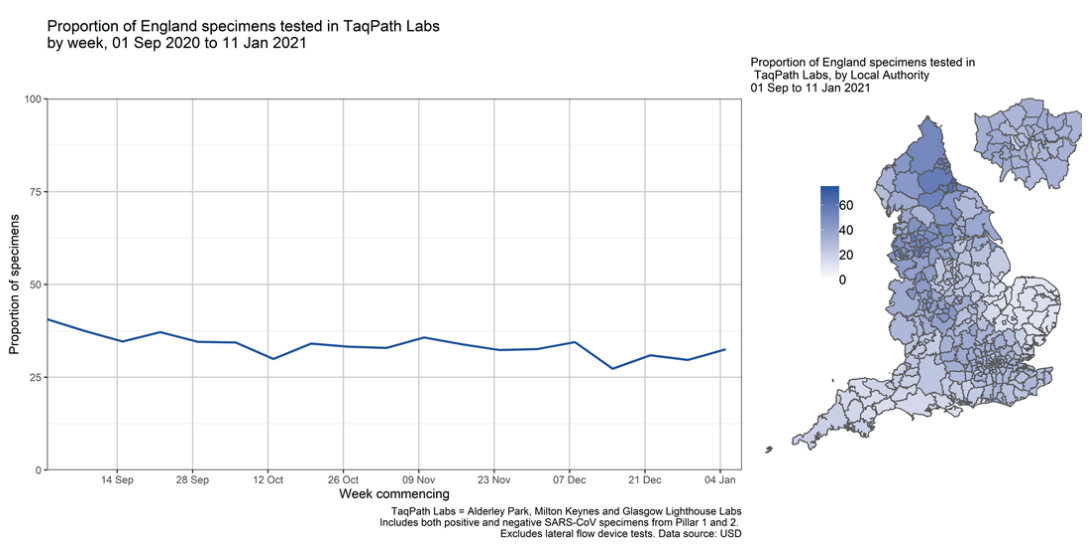
Figure 4. Proportion of Pillar 2 COVID-19 cases with SGTF among those tested in TaqPath Labs, by local authority(17 November 2020 to 11 January 2021)
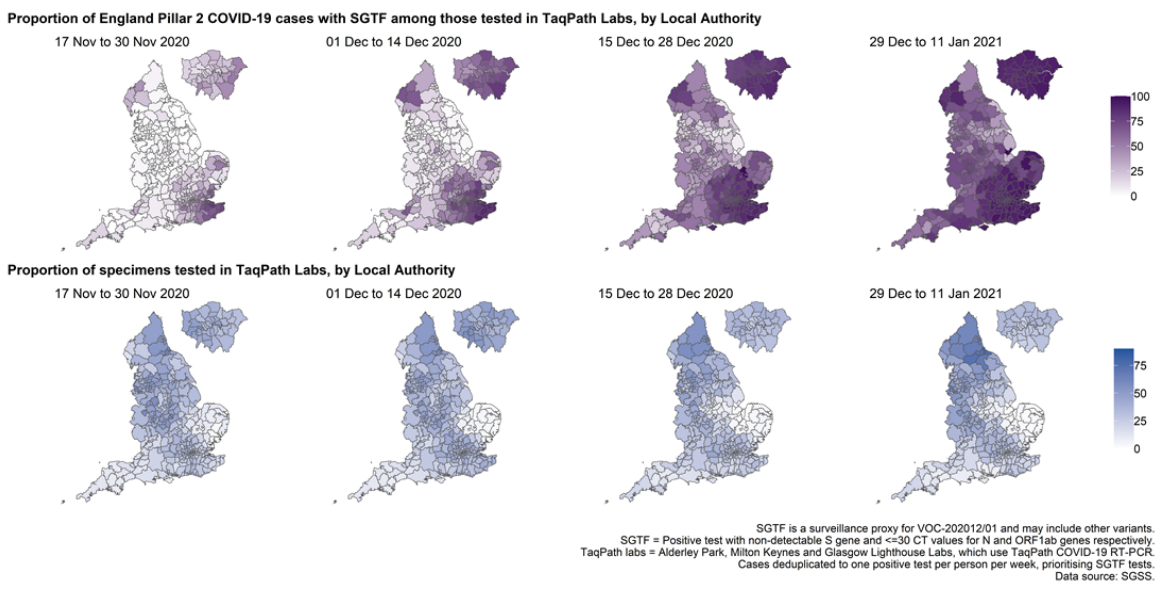
Figure 5. Weekly number (bars) and proportion (red lines) of Pillar 2 cases tested by TaqPath labs, by S-gene detection(1 September 2020 to 11 January 2021)
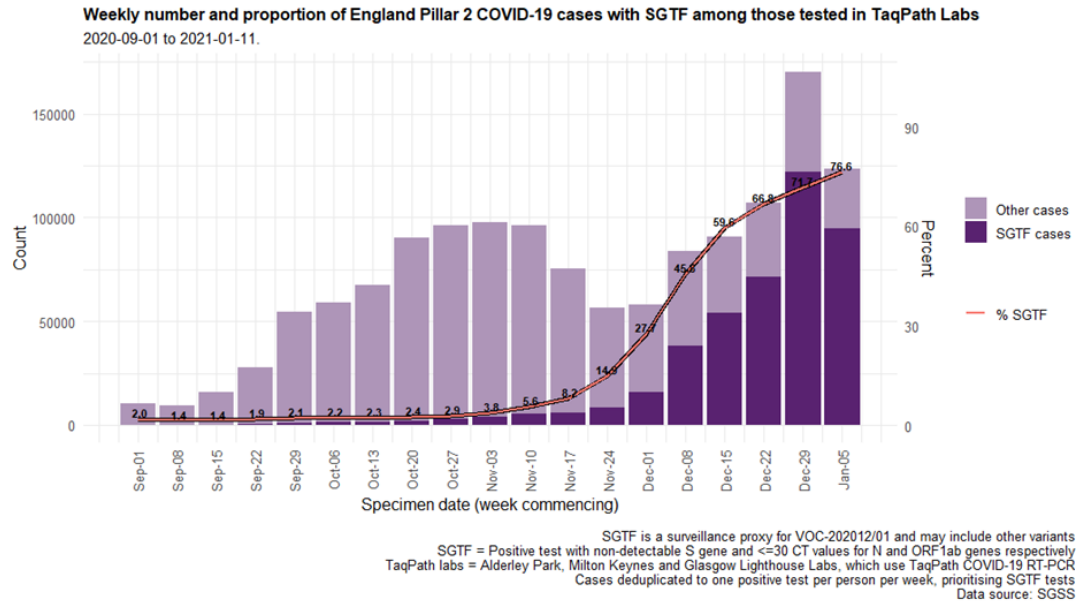
Figure 6. Weekly number (bars) and proportion (red lines) of Pillar 2 cases tested by TaqPath labs, by S-gene detection and region (1 September 2020 to 11 January 2021)
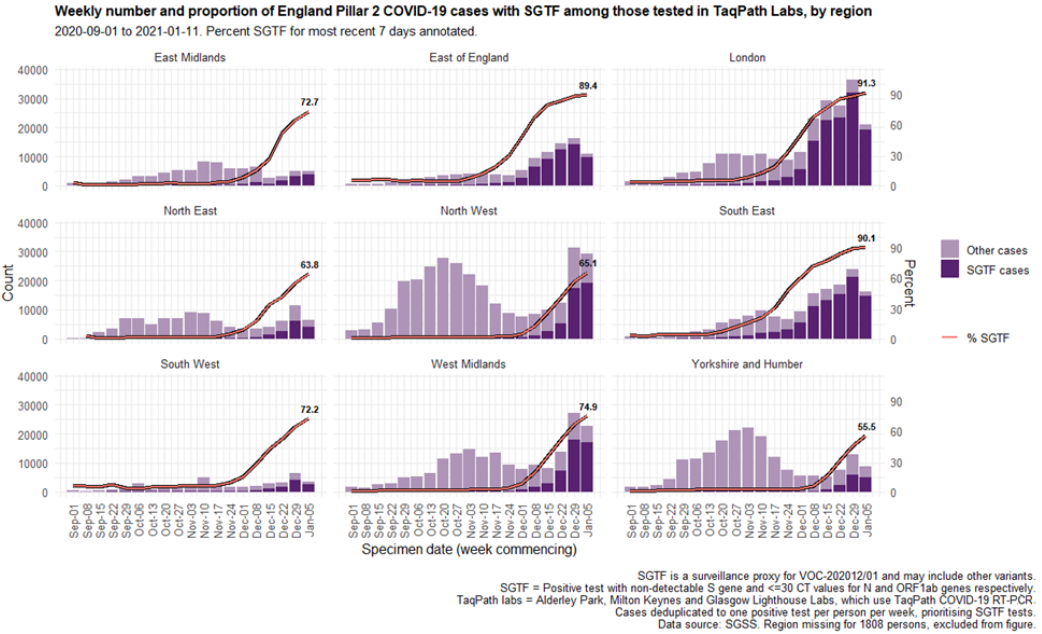
Figure 7. Sex-age pyramid of COVID-19 cases tested by TaqPath labs, S-gene detection, Pillar 2 cases only (1 December 2020 to 11 January 2021)
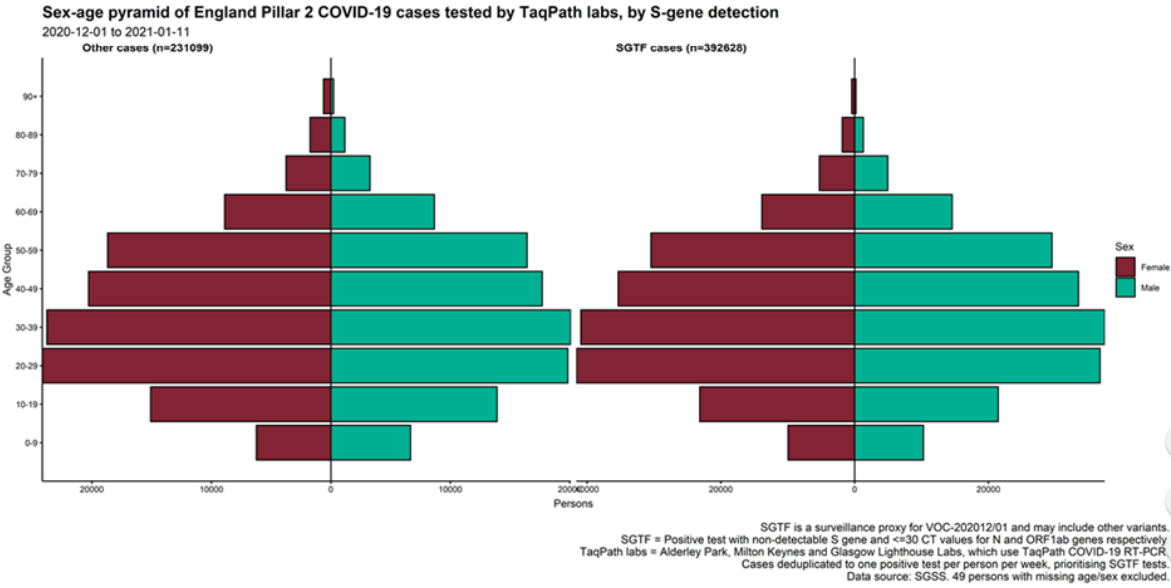
Figure 8. Weekly number (bars) and proportion (red lines) of Pillar 2 cases tested by TaqPath labs, by S-gene detection and age group (1 September 2020 to 11 January 2021)
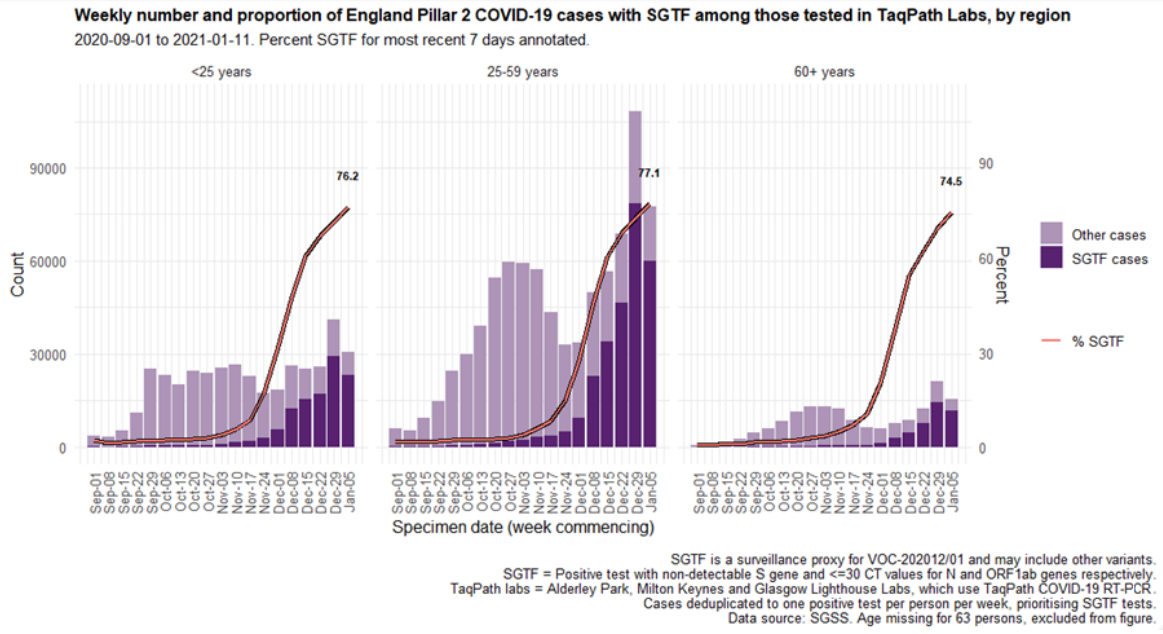
Office of National Statistics data
Office National Statistics Coronavirus (COVID-19) Infection Survey data is regularly updated
and available here including estimates of COVID-19 cases to 02 January for England, regions of
England and by cases compatible with the new variant (VOC 202012/01).
Summary
There is consistent evidence of cross-neutralising activity in convalescent sera (sera from individuals who have been infected with B1.1.7 shows neutralising activity against virus from other lineages, and the converse is also true).
Data sources
Data used in this investigation is derived from the COG-UK dataset, the PHE Second
Generation Surveillance System, NHS Test and Trace, the secondary uses service (SUS)
dataset and Emergency Care Data Set (ECDS).
GISAID reference genome
Sequences from this VOC can be identified by searching for the B1.1.7 lineage on
GISAID (gisaid.org). The canonical VOC genome is deposited with accession
EPI_ISL_601443.
Authors:Variant technical group
PHE: Meera Chand, Susan Hopkins, Christina Achison, Charlotte Anderson, Hester
Allen, Paula Blomquist, Cong Chen, Vicki Chalker, Gavin Dabrera, Obaghe Edeghere,
Matt Edmunds, Theresa Lamagni, Richard Myers, Isabel Oliver, Richard Elson, Eileen
Gallagher, Natalie Groves, Gareth Hughes, Meaghan Kall, Hannah Moore, Will Sopwith,
Charlie Turner, Lara Utsi, Marina Vabistsevits, Roberto Vivancos, Asad Zaidi, Maria Zambon;
Imperial College London:Wendy Barclay, Neil Ferguson, Erik Volz;
University of Birmingham:Nick Loman;
University of Edinburgh:Andrew Rambaut;
Wellcome Sanger Institute:Jeff Barrett
Twitter: @PHE_uk
Acknowledgements: The authors are grateful to those teams and groups providing
data for this analysis, including the Lighthouse Laboratories, COG-UK, the Wellcome
Sanger Institute, the PHE Contact Tracing Cell, Epidemiology Cell, Genomics Cell
and Outbreak Surveillance team.
For queries relating to this document, please contact: [email protected]
Version 1, release date 5 January 2021

Published January 2021
PHE gateway number: GW-1856






