Investigation of novel SARS-CoV-2 variant
Variant of Concern 202012/01
Technical briefing 5
This briefing provides an update on the briefing of 14 January 2021
Summary
SGTF detections indicate VOC202012/01 continues to predominate across all regions. S gene
target failure remains a well correlated proxy measure of VOC 202012/01. An assessment of
severity of disease has been conducted by NERVTAG. A limited number of B.1.1.7 VOC
202012/01 genomes with E484K mutation have been detected.
Nomenclature of variants in the UK
SARS-CoV-2 variants if considered to have concerning epidemiological, immunological or
pathogenic properties are raised for formal investigation. At this point they are designated
Variant Under Investigation (VUI) with a year, month, and number. Following risk assessment
with the relevant expert committee, they may be designated Variant of Concern (VOC). This
variant was designated VUI 202012/01 on detection and on review re-designated as VOC
202012/01 on 18 December 2020.
Severity of disease
TA summary of the current data and analyses on severity of disease associated with B.1.1.7 is
available at:
https://www.gov.uk/government/publications/nervtag-paper-on-covid-19-variant-of-concern-b117
Epidemiological findings
Updated 26 January 2021 to include new data. A matched cohort study was undertaken with
SGTF and non-SGTF cases, with matching based on 10-year age bands, sex, week of test and
lower-tier local authority. There were 92,207 SGTF cases and an equal number of
corresponding comparators included in the matched cohort (n = 184,414). A 28 day case fatality
risk was calculated for both SGTF and non-SGTF cases. Initial analysis was completed on 8
January 2021; of 14,939 SGTF cases and 15,555 comparators who had at least 28 days
between specimen date and the study period end date. There were 25 deaths (0.17%) in SGTF
cases and 26 deaths (0.17%) in comparators (RR 1.00, 95% CI 0.58 – 1.73). On 19/01/2021,
updated linkage of deaths data to the same matched cohort provided increased time for followup
and ascertainment of deaths identified there were 65 deaths among non-SGTF cases (0.1%)
and 104 deaths among SGTF cases (0.2%), within 28 days of specimen date. With this, the risk
ratio increased to 1.65 (95%CI 1.21-2.25).
S gene target failure/lineage correlation
Only a small fraction of all new cases of VOC 202012/01 are identified by whole-genome
sequencing, and this data typically lags test date by approximately 2 weeks, therefore a proxy S
gene target failure (SGTF) is used to monitor the VOC.
We previously observed that one of the S gene mutations in the VOC, which deletes amino
acids 69 and 70 (Δ69-70), causes a reproducible S gene target failure (SGTF) in the
ThermoFisher TaqPath assay used in 3 UK lighthouse laboratories (see Technical Briefing 1).
These laboratories are referred to here as ‘TaqPath laboratories.’ The Lighthouse laboratories
provide testing for samples from the community.
This coincidental occurrence provides a good proxy for monitoring trends in VOC 202012/01.
SGTF correlates almost perfectly with presence of Δ69-70. Considering 31,284 tested pillar 2
samples where we know both the sequence and the SGTF status, 99.6% of Δ69-70 sequences
(12,675 of 12,720) are SGTF, compared to 0.05% of sequences without the deletion (9 of
18,564).
Because Δ69-70 has arisen multiple times, and SGTF is a proxy for any lineage with that
mutation, the utility of SGTF as a proxy for VOC 202012/01 varies over time and region. Table 1
shows, for all pillar 2 sequences, the weekly proportion of Δ69-70 sequences that were
confirmed to be VOC 202012/01. Table 2 shows the proportion of Δ69-70 that is the VOC
202012/01 in England since December 21, broken down by region. It is now over 99% in all
regions of England. The numbers in these tables are based on sequenced samples, some of
which may have come from the same individual (this effect is likely to be small).
| Week beginning | Per cent VOC of all Δ69-70 | Number of pillar 2 Δ69-70 sequences |
| 2020-10-12 | 3% | 116 |
| 2020-10-19 | 15% | 220 |
| 2020-10-26 | 29% | 156 |
| 2020-11-02 | 64% | 399 |
| 2020-11-09 | 81% | 711 |
| 2020-11-16 | 89% | 771 |
| 2020-11-23 | 93% | 387 |
| 2020-11-30 | 95% | 423 |
| 2020-12-07 | 98% | 2704 |
| 2020-12-14 | 99% | 4301 |
| 2020-12-21 | 99% | 2400 |
| 2020-12-28 | 99.7% | 4766 |
| 2021-01-04 | 99.7% | 4509 |
| 2021-01-11 | 99.9% | 972 |
| Region | Percent VOC 202012/01 of all Δ69-70 | Number of pillar 2 Δ69-70 1 December to 18 January |
| East Midlands | 99.6% | 282 |
| East of England | 99.7% | 1567 |
| London | 99.7% | 3845 |
| North East | 99.4% | 470 |
| North West | 99.6% | 1808 |
| South East | 99.5% | 1908 |
| South West | 100% | 329 |
| West Midlands | 99.2% | 1320 |
| Yorkshire and the Humber | 100% | 430 |
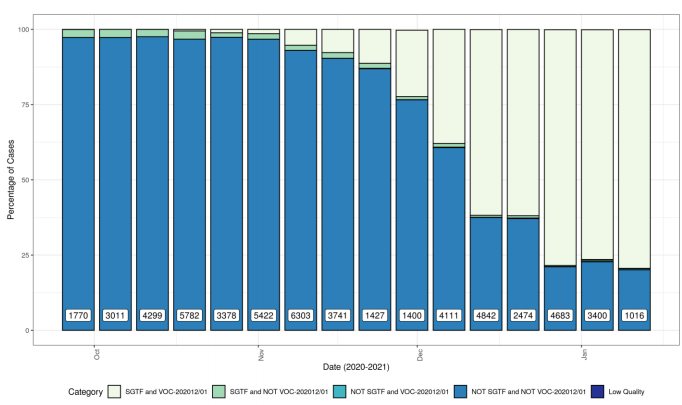
Figure 1 shows the weekly percentage of VOC202012/01 and STGF combinations, showing
increasing proportion of VOC202012/01 with SGTF (Pillar 2 samples with CT <30).
In addition there is now a small amount of data from hospital settings (unpublished).
A study of 82 sequences was undertaken in a single hospital in January 2021. 100% (66/66)
SGTF samples were confirmed as VOC 202012/01 by sequencing. 15/15 non VOC 202012/01
genomes had S gene ampification. A single VOC 202012/01 genome that was S gene positive
demonstrated S gene amplifcation >5 CT compared to N and ORF1ab results on multiplex
testing of the same sample (TaqPath PCR). Diagnostic labs should be aware of the possibility
of late S gene amplification as another potential presentation of variant VOC 202012/01,
although additional genomic confirmation studies are required.
A second study included 636 samples at two hospitals tested by the Thermofisher TaqPath
PCR over weeks 44-53 2020. Those samples with a CT of <30 in at least one target were
sequenced. Analysis by week shows that the VOC 202012/01 is the predominant cause of
SGTF by week 50 and was the cause of almost all SGTF in week 52/53. Lineage B. 1.258 is
noted as a cause of SGTF in earlier samples and individual hospitals may be affected by local outbreaks, altering circulating lineages. A small number of ‘late’ S gene target positives in VOC 202012/01 samples, as highlighted above were also identified in this study.
Diagnostic labs should be aware of the possibility of late S gene amplification as another
potential presentation of VOC 202012/01, although additional genomic confirmation studies are
required.
Epidemiology of S gene target failure
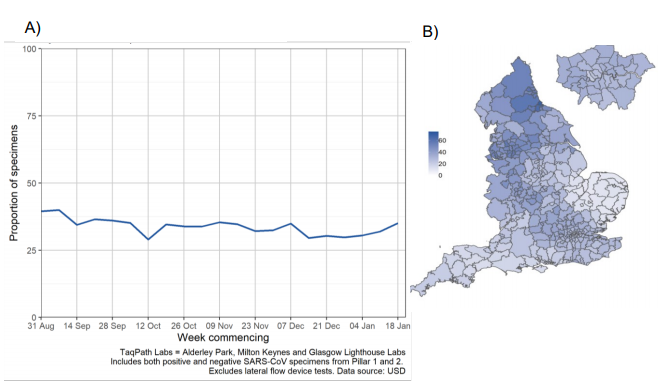
Since 1 September 2020, the proportion of England specimens tested in TaqPath laboratories
has remained at approximately 35% (Figure 2). This coverage is however lower in local
authorities in the East and South West of England.
Descriptive analyses below are restricted to TaqPath lab Pillar 2 positive tests with CT values
<=30 for non S gene targets. This restriction to CT values removes potential confounders
around variable target performance at lower viral loads, and is consistent with the sample set for
which genomic confirmation is available.
The following definitions apply (please note the change from previous reports):
• confirmed SGTF: Positive test with non-detectable S gene and <=30 CT values for N
• confirmed S gene positive: Positive test with <=30 CT values for S, N, and ORF1ab genes
In the most recent 7-day period (18 to 24 January 2021), 89.5% of 75,092 cases with S gene
detection results had isolates with SGTF, compared to 85.8% in the prior seven day period and
28.2% in the seven day period starting November 30th (Figure 3). This increase coincides with
the overall case number increase across England, however has persisted in recent weeks while
case counts decrease.
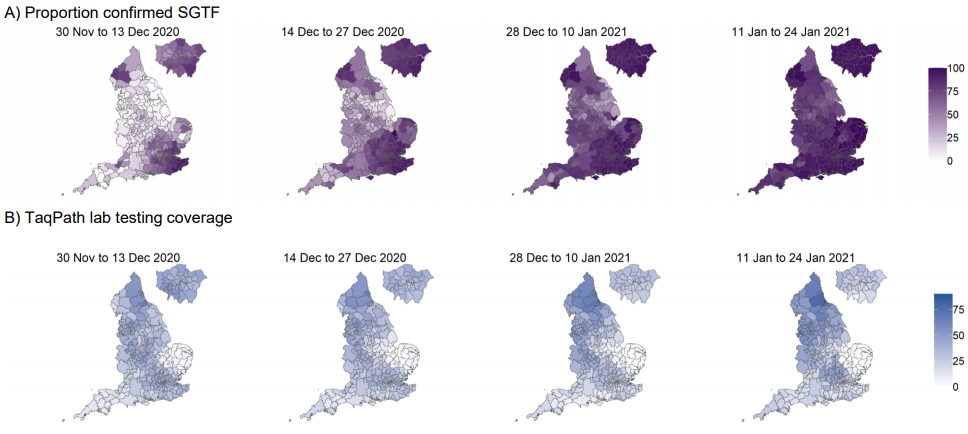
By region, the proportion of cases with SGTF has reached nearly 100% in London, the South
East, and East of England (Figure 4), whereas other areas see lower proportions that continue
to rise. This geographic spread from these areas to the rest of England is evident in Figure 5,
although higher proportions of SGTF were also detected earlier on in some local authorities of
the North West.
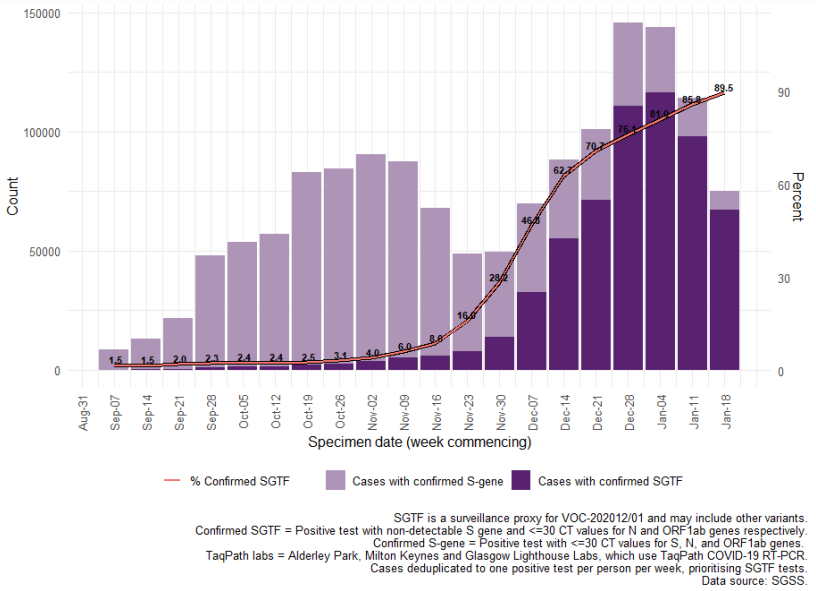
Figure 4. Weekly number (bars) and proportion (red lines) of Pillar 2 COVID-19 cases tested by TaqPath laboratories with SGTF among those with S gene detection results, by region of residence (7 September 2020 to 24 January 2021). Percent confirmed SGTF for most recent days annotated.
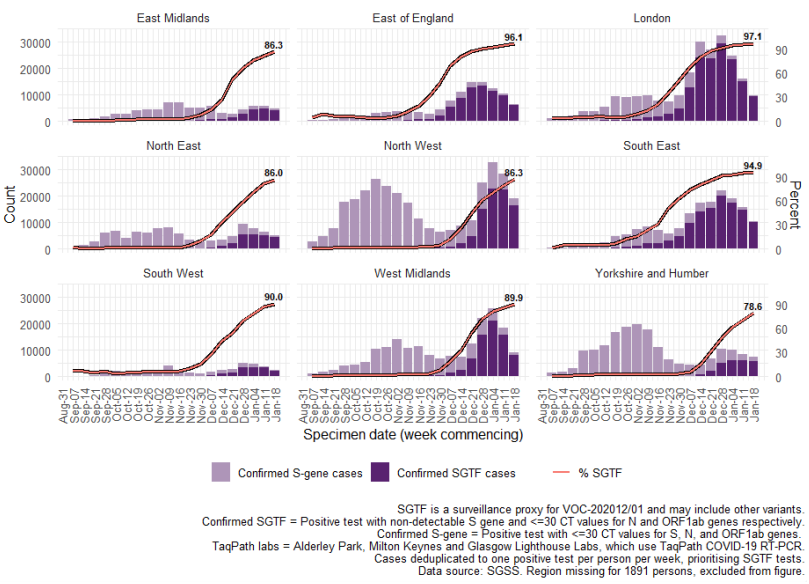
Figure 5. Local authority maps of A) proportion of Pillar 2 COVID-19 cases with SGTF among those tested in TaqPath laboratories and with S gene detection results, and B) Proportion of all specimens tested in TaqPath laboratories (30 November 2020 to 24 January 2021)
Cases by age and sex are displayed (Table 3). Proportions of cases with SGTF are highest in cases aged 10 or younger, at 74.1%
of 44,627 cases between 1 December 2020 and 24 January 2021, and lowest in cases aged 75 or older, at 64.9% of 19,216 cases
within the same period.
| Females | Males | Total | |||||||
| Age group | Cases | SGTF cases | Percent SGTF | Cases | SGTF cases | Percent SGTF | Cases | SGTF cases | Percent SGTF |
| ≤10 years | 21716 | 16145 | 74.3 | 22911 | 16945 | 74.0 | 44627 | 33090 | 74.1 |
| 11-16 years | 23872 | 16526 | 69.2 | 22686 | 15605 | 68.8 | 46558 | 32131 | 69.0 |
| 17-24 years | 55307 | 40237 | 72.8 | 46199 | 34102 | 73.8 | 101507 | 74339 | 73.2 |
| 25-59 years | 252278 | 182272 | 72.3 | 231789 | 170595 | 73.6 | 484071 | 352870 | 72.9 |
| 60-74 years | 36215 | 25374 | 70.1 | 36811 | 26340 | 71.6 | 73026 | 51714 | 70.8 |
| 75+ years | 11384 | 7381 | 64.8 | 7832 | 5096 | 65.1 | 19216 | 12477 | 64.9 |
| Total | 400796 | 287952 | 71.8 | 368261 | 268709 | 73.0 | 769062 | 556664 | 72.4 |
Data on coverage of TaqPath laboratories testing and numbers/proportions of cases with SGTF are shared daily with Local
Authorities (Sunday-Friday) on the COVID-19 PHE Local Authorities Report Store (Sharepoint).
Analysis of secondary attack rates using routine contact tracing data
This data has been updated from the previous briefing to include cases reported up to the
10 January 2021 (analysed on 26 January 2021). Findings are similar to those previously
reported(www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variantvariant-of-concern-20201201).
PHE have analysed secondary attack rates among contact tracing data (from NHS Test
and Trace) for the variant of concern (VOC 202012/01) using both genomic sequence
variant data and S gene target failure (SGTF) data for Pillar 2 cases with valid S gene
detection results, as defined above.
Between 30 November 2020 and 10 January 2021, 1,364,301 cases were reported to
NHS Test and Trace. 35,597 (2.6%) had genomic sequencing data included*; 18,160
(51%) of those cases had a result of confirmed VOC 202012/01. 572,356 (42%) had valid
S gene detection results; 376,274 (65.7%) of those cases had SGTF.
2,722,845 contacts reported to NHS Test and Trace were identified between 30 November
2020 and 10 January 2021. 66,847 contacts were reported by cases with genomic
sequencing data; 37,585 of those contacts were reported by cases with VOC 202012/01.
In all, 46.5% (1,266,461) Pillar 2 contacts of cases who were tested for S gene; of which
68.4% (866,608) had SGTF.
308,038 (11.3%) of all contacts were infected (secondary attack rate)
• 12.9% among those whose index case with genomic data had confirmed VOC
202012/01. 12.9% among those whose index case had laboratory detections of SGTF.
• 9.7% among those whose index case had a genomic result of wild type. 9.9%
among those whose index case had confirmed presence or absence of S gene and did not
have SGTF.
Both when using genomic sequence data directly and SGTF as a proxy, the secondary
attack rates estimated from contact tracing data were observed to be higher if the index
case has the variant strain, from around 10% to 13% of named contacts. This increase
was around 10%-55% across most age groups and regions where sufficient sequencing
data is available. Using the SGTF proxy to give a more comprehensive overview the
increase was consistently around 25%-40%.
Attack rate: contacts becoming cases
| Characteristic of contact | Allcontacts | Contacts of people with VOC 202012/01 | Contacts of people with wild type (not VOC 202012/01) | |||||
| Total contact | All contacts | Contacts that became cases | % | All contacts | Contacts that became cases | % | ||
| Region of residence | All | 2,722,845 | 37,585 | 4,845 | 12.9 | 24,239 | 2,340 | 9.7 |
| East Midlands | 172,594 | 868 | 100 | 11.5 | 1,734 | 185 | 10.7 | |
| East of England | 406,849 | 6,801 | 876 | 12.9 | 2,358 | 243 | 10.3 | |
| London | 721,377 | 12,883 | 1,624 | 12.6 | 3,464 | 299 | 8.6 | |
| North East | 85,273 | 1,104 | 122 | 11.1 | 1,795 | 178 | 9.9 | |
| North West | 274,688 | 3,577 | 542 | 15.2 | 5,121 | 495 | 9.7 | |
| South East | 496,778 | 7,503 | 912 | 12.2 | 2,044 | 167 | 8.2 | |
| South West | 150,985 | 1,011 | 131 | 13.0 | 1,459 | 160 | 11.0 | |
| West Midlands | 259,729 | 2,806 | 392 | 14.0 | 2,891 | 279 | 9.7 | |
| Yorkshire and Humber | 146,304 | 949 | 130 | 13.7 | 3,301 | 329 | 10.0 | |
| Level of contact* | Direct | 2,513,595 | 34,674 | 4,636 | 13.4 | 21,777 | 2,215 | 10.2 |
| Close | 200,131 | 2,712 | 202 | 7.4 | 1,964 | 114 | 5.8 | |
| Age group | 0–9 | 381,835 | 5,306 | 427 | 8.0 | 3,242 | 172 | 5.3 |
| 10–19 | 455,374 | 6,463 | 668 | 10.3 | 4,064 | 349 | 8.6 | |
| 20–29 | 357,008 | 4,792 | 711 | 14.8 | 3,016 | 367 | 12.2 | |
| 30–39 | 317,666 | 4,393 | 754 | 17.2 | 2,820 | 309 | 11.0 | |
| 40–49 | 327,015 | 4,575 | 725 | 15.8 | 2,897 | 355 | 12.3 | |
| 50–59 | 304,303 | 4,149 | 740 | 17.8 | 2,691 | 358 | 13.3 | |
| 60–69 | 142,712 | 1,892 | 349 | 18.4 | 1,377 | 180 | 13.1 | |
| 70–79 | 55,456 | 708 | 157 | 22.2 | 550 | 111 | 20.2 | |
| 80+ | 23,102 | 273 | 55 | 20.1 | 250 | 36 | 14.4 | |
| Not known | 358,374 | 5,034 | 259 | 5.1 | 3,332 | 103 | 3.1 | |
| Characteristic of contact | Allcontacts | Contacts of people with S-gene target failure | Contacts of people with wild type (no S-gene target failure) | |||||
| All | All contacts | Contacts that became cases | % | All contacts | Contacts that became cases | % | ||
| Region of residence | ||||||||
| East Midlands | 172,594 | 25,988 | 3,373 | 13.0 | 32,548 | 3,128 | 9.6 | |
| East of England | 406,849 | 117,176 | 15,113 | 12.9 | 23,471 | 2,376 | 10.1 | |
| London | 721,377 | 237,141 | 28,311 | 11.9 | 46,290 | 4,270 | 9.2 | |
| North East | 85,273 | 33,739 | 4,667 | 13.8 | 31,662 | 3,241 | 10.2 | |
| North West | 274,688 | 103,121 | 14,320 | 13.9 | 89,440 | 9,420 | 10.5 | |
| South East | 496,778 | 179,693 | 23,124 | 12.9 | 33,834 | 3,366 | 9.9 | |
| South West | 150,985 | 23,366 | 3,179 | 13.6 | 15,617 | 1,571 | 10.1 | |
| West Midlands | 259,729 | 110,585 | 14,662 | 13.3 | 72,559 | 7,031 | 9.7 | |
| Yorkshire and Humber | 146,304 | 33,208 | 4,477 | 13.5 | 53,341 | 5,198 | 9.7 | |
| Level of contact* | Direct | 2,513,595 | 801,510 | 106,350 | 13.3 | 364,585 | 37,591 | 10.3 |
| Close | 200,131 | 62,733 | 5,133 | 8.2 | 33,288 | 2,073 | 6.2 | |
| Age group | 0–9 | 381,835 | 123,487 | 9,366 | 7.6 | 56,639 | 3,043 | 5.4 |
| 10–19 | 455,374 | 147,357 | 15,843 | 10.8 | 68,885 | 5,941 | 8.6 | |
| 20–29 | 357,008 | 114,669 | 17,477 | 15.2 | 49,838 | 6,119 | 12.3 | |
| 30–39 | 317,666 | 100,478 | 16,396 | 16.3 | 46,773 | 5,910 | 12.6 | |
| 40–49 | 327,015 | 105,153 | 17,039 | 16.2 | 48,889 | 5,964 | 12.2 | |
| 50–59 | 304,303 | 97,345 | 16,946 | 17.4 | 45,493 | 5,990 | 13.2 | |
| 60–69 | 142,712 | 43,780 | 8,019 | 18.3 | 21,177 | 2,955 | 14.0 | |
| 70–79 | 55,456 | 15,933 | 3,142 | 19.7 | 8,037 | 1,212 | 15.1 | |
| 80+ | 23,102 | 6,259 | 1,173 | 18.7 | 3,317 | 542 | 16.3 | |
| Notknown | 358,374 | 112,147 | 6,120 | 5.5 | 50,805 | 2,014 | 4.0 | |
*Direct: face to face contact (e.g. a conversation within 1 metre); skin to skin contact
(including sexual contact); coughed on, sneezed on or spat on. Close: within 1 metre for 1
min. or more (not necessarily face to face); within 1-2 metres for 15 mins. or more (could
be total 15 mins. over 24 hours); travelling in a small vehicle; travelling in a large vehicle or
plane (1 metre for 1 min. and 1-2 metres for 15 mins.).
Estimated attack rates for cases with VOC 202012/01 are 10%-55% higher than estimated
attack rates with sequencing and other variants/ lineages for most regions and age groups,
excepting the East Midlands (which has relatively small numbers of people with genomic
results).
Estimated attack rates for cases with SGTF are 25% - 40% higher than estimated attack rates for cases with S gene detection results and no SGTF for most regions and age groups, excepting groups with few records such as the 80+ age group.
Detection of E484K mutation in B.1.1.7 VOC 202012/01
The COG-UK dataset (total sequences 214,159) was analysed on 26/01/2021. The spike protein mutation E484K (found in VOC 202012/02 B1.351 and VOC 202101/02 P1) has been detected in 11 B1.1.7 sequences. Preliminary information suggests more than one acquisition event.
Office of National Statistics data
Office National Statistics Coronavirus (COVID-19) Infection Survey data is regularly
updated and available here including estimates of COVID-19 cases for England, regions of
England and by cases compatible with the new variant (VOC 202012/01).
It includes new symptom comparison data:
http://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/articles/coronaviruscovid19infectionsinthecommunityinengland/characteristicsofpeopletestingpositiveforcovid19in
Data sources
Data used in this investigation is derived from the COG-UK dataset, the PHE Second
Generation Surveillance System, NHS Test and Trace, the secondary uses service (SUS)
dataset and Emergency Care Data Set (ECDS).
GISAID reference genome
Sequences from this VOC can be identified by searching for the B1.1.7 lineage on
GISAID (gisaid.org). The canonical VOC genome is deposited with accession
EPI_ISL_601443.
Contact: All enquiries relating to scientific or public health matters should be addressed to
[email protected]
Variant Technical Group
This group includes representation with the following organisaions:
PHE, DHSC, BEIS, Wales NHS, PHScotland, NHS Scotland, Health and Social Care
Northern Ireland
Imperial College London, London School of Hygiene & Tropical Medicine, University of
Birmingham, University of Cambridge, University of Edinburgh, University of Liverpool.
Wellcome Sanger Institute
Additional contributions were received from
The COG-UK-HOCI study, University College London: Judith Breuer
University Hospitals Birmingham NHS Foundation Trust
Acknowledgements: The authors are grateful to those teams and groups providing data for
this analysis, including the Lighthouse Laboratories, COG-UK, the Wellcome Sanger
Institute, the PHE Epidemiology Cell, Contact Tracing, Genomics and Outbreak
Surveillance Teams.

Published January 2021
PHE gateway number: GW-1856






