The strategies used in diagnosis of human Infectious diseases
Abstract
Infectious diseases are a significant burden on public health and economic stability of societies all over the world. They have been among the leading causes of death and disability and presented growing challenges to health security and human progress for centuries. Infectious diseases are generally caused by microorganisms. The routes of them entry into host is mostly by the mouth, eyes, genital openings, nose, and the skin. Damage to tissues mainly results from the growth and metabolic processes of infectious agents intracellular or within body fluids, with the production and release of toxins or enzymes that interfere with the normal functions of organs and/or systems [1]. Advances in basic science research and development of molecular technology and diagnostics have enhanced understanding of disease etiology, pathogenesis, and molecular epidemiology, which provide basis for appropriate detection, prevention, and control measures as well as rational design of vaccine [2]. The diagnosis of infectious diseases is particularly critical for the prevention and control of the epidemic. Here we introduce the insights and detection methods of infectious disease, aiming to provide some helps for clinical diagnosis as well as epidemic prevention and control of infectious diseases.The strategies used in diagnosis of human Infectious diseases
1 Molecular Methods
The development of molecular methods for the direct identification of a specific viral genome from the clinical sample is one of the greatest achievements of the 21st century. Clearly nucleic acid amplification techniques including Reverse Transcription-Polymerase Chain Reaction (RT-PCR), nucleic acid sequence-based amplification (NASBA) and Lawrence Livermore Microbial Detection Array (LMDA) are proven technology leaders for rapid detection and molecular identification for most known human viruses [9].
RT-PCR assays for virus detection provides faster results than end-point assays and in many cases have sensitivities equal to or better than culture [10]. The novel coronavirus, 2019-nCoV, was detected through real-time RT-PCR with primers against two segments of its RNA genome [11]. The particular primer sets and specific guideline for detection of COVID-19 through RT-PCR were made available by the Center for Disease Control (CDC) USA, according to CDC [12]. However, high mutation rates may lead to extensive changes in viral nucleic acid sequences making dedicated PCR primer use irrelevant, therefore there is high demand for the development of rapid and universal virus identification and detection technologies. In contrast, although NASBA assay is considered sensitive; it has not been widely used because of the difficulties in the preparation of NASBA master mix in-house and the high cost of commercial kits. A new molecular biology-based microbial detection method for rapid identification of multiple virus types in the same sample has been developed by a research group at Lawrence Livermore National Laboratory. Lawrence Livermore Microbial Detection Array (LLMDA) detects viruses using probes against genomic DNA sequence within 24 hours [13,14]. In addition, the oligonucleotide probes were selected to enable detection of novel, divergent species with homology to sequenced organisms [14].
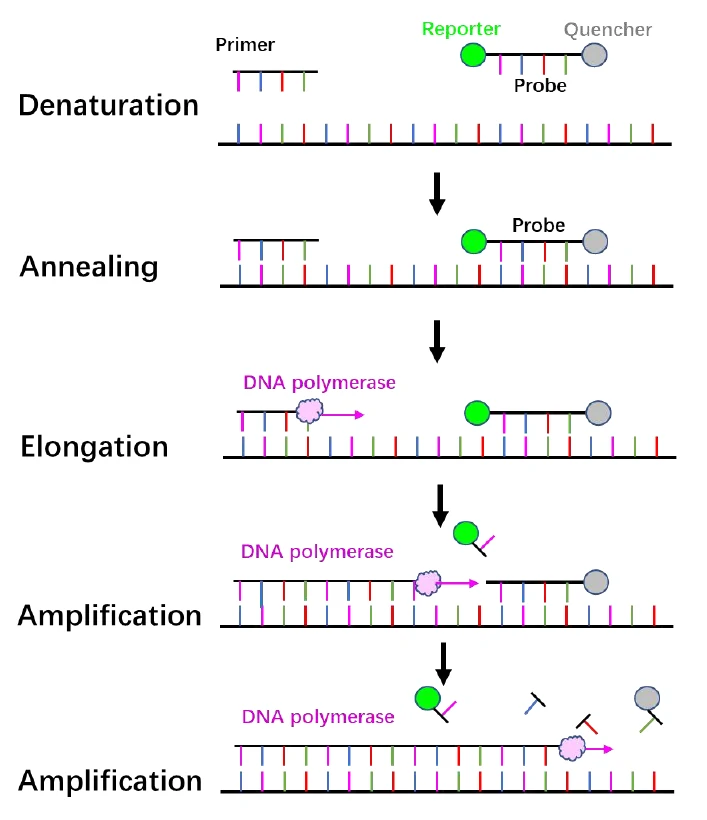 Figure 1. Principle of RT-PCR.
Figure 1. Principle of RT-PCR.
2 Immuno-assays
The nucleic acid diagnostic tool currently employed is with good sensitivity and excellent specificity. However, due to its high false negative, time-consuming, high level equipment and technical personnel demand, the immunological antigen or antibody detection has been paid more and more attention because of its quick detection speed, low and simple technical requirements of detection. At present, the detection methods mainly include Enzyme-linked immunosorbent assays (ELISAs), colloidal gold immunochromatography (GICA) and magnetic particle chemiluminescence.
2.1 Enzyme-Linked Immunosorbent Assays (ELISA)
Enzyme-linked immunosorbent assays (ELISAs) incorporate the sensitivity of simple enzyme assays with the specificity of antibodies, by employing antigens or antibodies coupled to an easily-assayed enzyme. As such ELISA is much more rapid method than immunoblotting to detect specific viral protein from a cell, tissue, organ, or body fluid. There are two main variations of ELISAs: antigen-capture ELISA (detecting viral proteins), involve attachment of a capture antibody to a solid matrix for the viral protein of interest, while antibody-capture ELISA measures the specific antibody level in a sample, by coating viral antigen protein on a solid surface. There are two principles based on antigen-capture and antibody-capture ELISAs. In a general, ELISAs are considered a highly sensitive method that can detect a fairly low number of proteins at the range of picomolar to nanomolar range (10-12 to 10-9 moles per liter). ELISA has been one of the most widely used serologic tests for detecting antibody to HIV-1. ELISA method was found useful as a diagnostic tool to detect influenza viral antigen much quicker than other conventional virus detection methods [15]. In another previous study, comparison of ELISA, with conventional methods has demonstrated ELISA superiority for the rapid detection and identification of influenza A virus [16]. A simplified and standardized neutralization enzyme immunoassay (Nt-EIA) was developed to detect measles virus growth in Vero cells and to quantify measles neutralizing antibody [17]. Newer EIA formats for hepatitis C virus diagnostics have been constantly evaluated [18,19]. As such ELISAs are being used for plethora of application both in experimental and diagnostic virology including HIV-1, dengue, and influenza [20-22]. On the other hand, although rapid than traditional plaque assays or TCID50, ELISA assays sometimes could be quite expensive, due to the cost of reagents used. Unfortunately, sometimes required antibodies may not be commercially developed as well. In contrast, attempts to develop antibodies in-house may be quite expensive. Additional variability may also be introduced due to high background signals generated by non-specific binding, or cross-reactivity with non-viral protein targets.
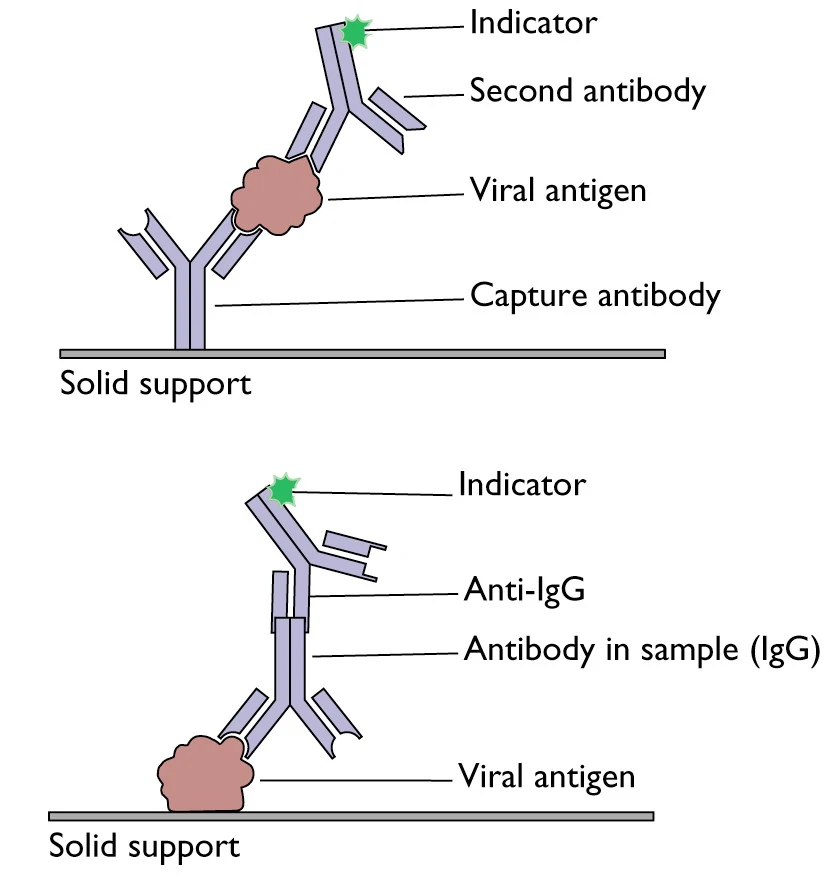 Figure 2. A schematic representation of two principles based on antigen or antibody capture ELISA[23]
2.2 Colloidal gold immunochromatography (GICA)
Figure 2. A schematic representation of two principles based on antigen or antibody capture ELISA[23]
2.2 Colloidal gold immunochromatography (GICA)Based on the specific immune response of antigen and antibody, colloidal gold particles were used as one of the tracer markers. Driven by solvent chromatography, the markers had an immune response on the C/T line, and the detection results could be obtained according to the color of the T line. GICA samples can be whole blood, serum or plasma, and studies have shown that the colloidal gold reagent has a high consistency in detecting whole blood, plasma or serum [24]. At present, there are seven kinds of colloidal gold kits approved by the State Food and Drug Administration, which are all detection antibodies, but there is no detection kit for antigens. RT-PCR was used as the control method, and the sensitivity and specificity of IgM/IgG antibody were different, and the highest detection rate of the two combined detection was 66.1% (125/189) [25]. This method can be used for enterprise resumption, students return to school, community crowd screening and other scenarios. Only a drop of fingertip blood is needed, and the detection results can be observed visually in 15 minutes, which is rapid and simple without special instruments. However, the detection has its disadvantages such as window period, without quantification, exposure risk, low sensitivity and vulnerability to environmental factors, and nucleic acid detection combination result is required for verification.
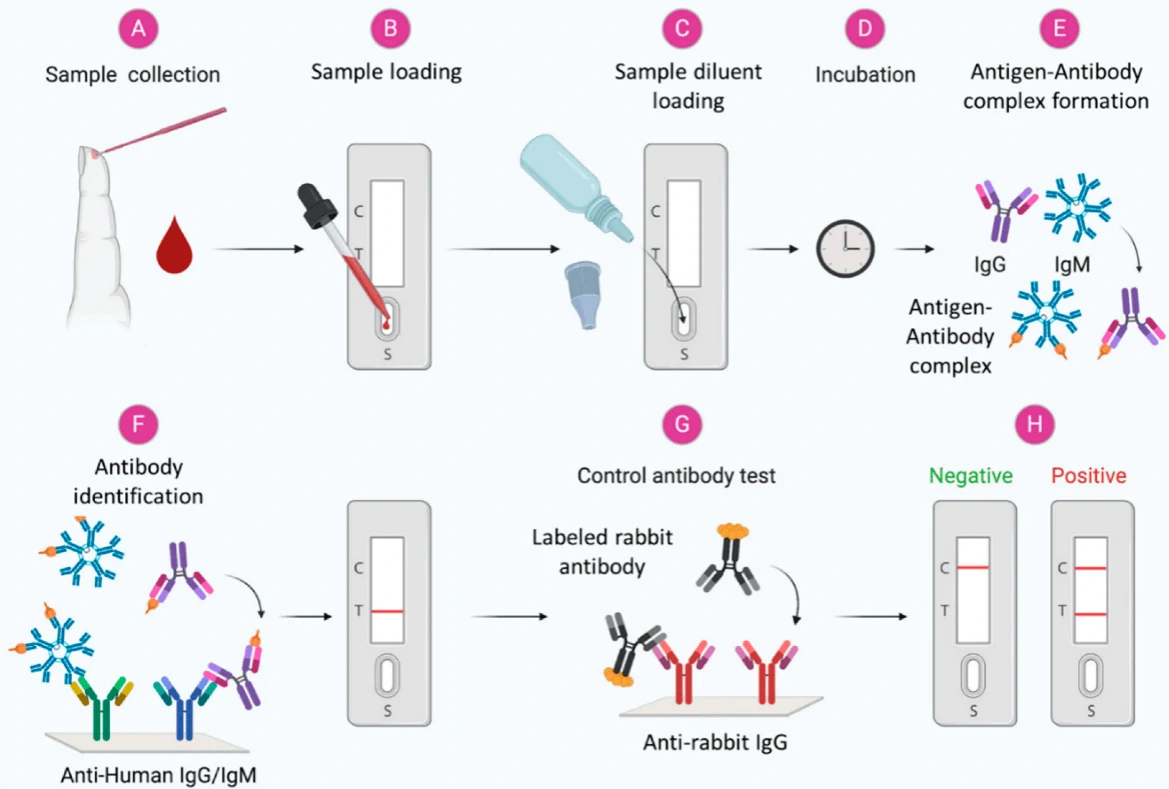 Figure 3. Scheme showing the general steps in the antibody-based diagnosis of viral infections from blood samples[26]
2.3 Magnetic particle chemiluminescence
Figure 3. Scheme showing the general steps in the antibody-based diagnosis of viral infections from blood samples[26]
2.3 Magnetic particle chemiluminescenceMagnetic particle chemiluminescence is an emerging technique to capture specific IgM/IgG antibodies in samples using magnetic particles fixed on the surface of recombinant antigens. The antigen-antibody complex was precipitated by external magnetic field, and the captured antigen-antibody complex was identified by enzyme-labeled secondary antibody, and the luminescent intensity was determined by chemiluminescence instrument after adding the luminescent agent, and then quantitative analysis was carried out. It is characterized by high sensitivity, high specificity and wide detection range, etc. There are currently seven approved magnetic particle chemiluminescence detection kits, the first one developed by Bioscience, with an automatic chemiluminescence analyzer, capable of detecting at a speed of 240 T/H with an initial reporting time of 30 min [27].
3 Viral Culture
Virus culture, isolation and identification are the gold standards for laboratory identification of pathogens. However, viral culture results do not yield timely results to inform clinical management. Shell-vial tissue culture results may take 1-3 days, while traditional tissue-cell viral culture results may take 3-10 days. Due to the long incubation time, high technical requirements, and must be carried out in a level III safe biological laboratory, it is not suitable for rapid virus diagnosis during the epidemic period [28].
4 Immunofluorescence (IF) Assay
Immunofluorescence (IF) technique is widely used for rapid detection of virus infections by identifying virus antigens in clinical specimens. IF staining is usually considered very rapid (about 1 to 2 hr) and overall gives a sensitive and specific viral identification [29-32]. Unfortunately, IF technique may not able to confirm the identity of all virus strains, for instance viruses of the “enterovirus” group; since most monoclonal antibodies (MAbs) for enteroviral identification have been shown to lack sensitivity, while cross-reactivity with rhinoviruses is extremely common [33]. In contrast, IF has been successfully used for better management of influenza virus infection and surveillance of influenza virus activity [30, 31]. As recommended by CDC, when influenza activity is low, positive results should be confirmed by direct immunofluorescence assay (DFA), viral culture, or RT-PCR, as false positive test results are more likely; while during peak influenza activity confirmatory testing using DFA, viral culture, or PCR must always be considered because a negative test may not rule out influenza viral infection. Interestingly, although IF is generally considered less sensitive then ELISA and PCR, a recent publication reports DFA as an optimal method for rapid identification of varicella-zoster virus (VZV), when compared with conventional cell culture [34]. In contrast, the Herpes simplex virus (HSV) DFA test accuracy was found very low (sensitivity 61%, specificity 99%), when tested to identify mucocutaneous HSV infection in children [35]. Furthermore, a monoclonal antibody designated CHA 437 was developed against HSV showed no cross-reactivity against the varicella-zoster virus, cytomegalovirus, or Epstein-Barr virus, however direct specimen testing resulted in overall low sensitivity (84.6%) and specificity (95.7%) [36]. On the other hand, an antigen detection assay for severe acute respiratory syndrome (SARS) coronavirus (CoV) could detect SARS-CoV in 11 out of 17 (65%) samples from SARS patients. As such IF technique is well-accepted laboratory diagnostics test, however, sometime these assays could be quite expensive, due to the cost of antibodies used. Additional variability may also be introduced due to non-specific binding, or cross-reactivity of commercially available antibodies [37].
As such IF technique is well-accepted laboratory diagnostics test, however, sometime these assays could be quite expensive, due to the cost of antibodies used. Additional variability may also be introduced due to non-specific binding, or cross-reactivity of commercially available antibodies.
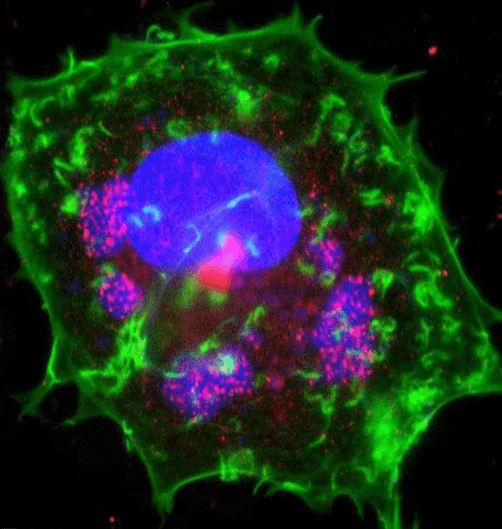 Figure 4. Immunofluorescence staining of vaccinia virus infected cell [38]. Areas of virus assembly within the cell are pink.
Figure 4. Immunofluorescence staining of vaccinia virus infected cell [38]. Areas of virus assembly within the cell are pink. Host and viral DNA (deoxyribonucleic acid) is blue. The host cell's DNA is contained within its nucleus (large oval). Actin protein
filaments, which make up part of the cytoskeleton, are green.
5 Immunoblotting (WB)
Immunoblotting technique detects specific viral proteins isolated from a cell, tissue, organ, or body fluid. The development of sensitive and specific tests for human immunodeficiency virus type 1 (HIV-1) progressed rapidly after this retrovirus was found to be responsible for causing AIDS [39]. Immunoblotting has been one of the reference confirmatory tests for the diagnosis of HIV infection or after inconclusive enzyme immunoassay (EIA) results. Although difficulty in interpretation of immunoblotting results and the cost led to a reduction in overall use of WB technique, nevertheless immunoblots are still commonly used for various purposes, including clinical diagnosis of HIV-1, seroprevalence surveys, and for blood-donor screening. In addition, immunoblot assays have been used to confirm the anti-hepatitis C virus (HCV) reactivity [40]. In recent years immunoblotting has been established as an important prerequisite for the functional studies to understand protein composition of the purified viral particles, since it allows the analysis of specific proteins which result in better understanding of the infection process and the pathogenesis of viruses [41,42].
6 Transmission Electron Microscopy (TEM)
Most viruses are very small to be seen directly under a light microscope, and therefore could only be viewed with TEM (transmission electron microscopy). In 1948, smallpox and chicken pox were first differentiated by TEM [43] and thereafter early virus classifications depended heavily on TEM analysis. In particular many intestinal viruses were discovered by negative staining TEM microscopy [44, 45]. Although TEM has gradually been replaced by more sensitive methods such as PCR, nevertheless it still remains essential for several aspects of virology including discovery, description and titration of viruses. One of the major advantages of using TEM is that it does not require virus-specific reagents; this is of particular importance in an outbreak setting where the etiologic agent is unknown and therefore specific reagents may not be available to determine correct detection tests. Negative stained TEM technique continues to be a valuable tool for the discovery and identification of novel viruses including Ebola virus, henipavirus (Hendra and Nipah) and SARS [46-50]. A human monkeypox outbreak was detected in the US by TEM [51]. Nevertheless, due to the high instrument cost and the amount of space and facilities required, TEM is still only available in certain facilities.
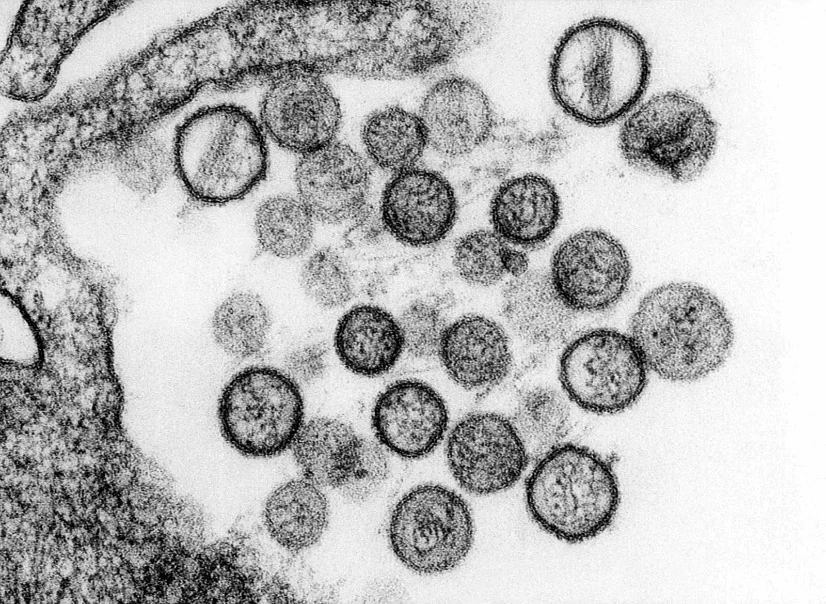 Figure 5. Transmission Electron Microscopy of hantavirus virions[52]
Figure 5. Transmission Electron Microscopy of hantavirus virions[52]Summary
Infectious diseases are a real public health threat, outbreaks can have serious social, political, and economic effects. A complex number of factors relating to human behavior and activities, pathogen evolution, poverty, and changes in the environment as well as dynamic human interactions with animals have been found to contribute to infectious disease emergence and transmission. Aggressive research is warranted to unravel important characteristics of pathogens necessary for diagnostics, therapeutics, and vaccine development. Here we describe some strategies for the diagnosis of human infectious diseases, hoping to be helpful for clinical diagnosis and epidemic prevention and control of infectious diseases. To date, multiple diagnostic techniques have been developed. Various diagnostic tools show both significances and limitations. Conventional approaches to quantify infective viral particles are labor-intensive, time-consuming, and often associated with poor reproducibility. Immunological tests generally provide quick results, however, is quite expensive due to the requirement of antigen-specific antibody. While RT-PCR may be able to provide results within a matter of hours, it is laborious, requires a skilled operator, and is sensitive to contamination. TEM-based quantification, although highly accurate in determining the shape and the total number of viral particles, often considered time-consuming, extremely expensive and impractical for high sample numbers. Moreover, TEM sample preparation is tedious, and the technique requires sophisticated instrument and a skilled operator. To alleviate these limitations, there is still a need to develop new cost-effective analytical methods that can allow users to quickly and easily determine virus concentrations and reduce constrictions coupled with current assays. Nevertheless, any such emerging methods must be carefully evaluated in terms of their efficiency, precision and linear range. The evaluation of each diagnostic technique and approval from the FDA are necessary before practical application.Products list: paired diagnostics grade antibodies (monoclonal antibody, mab) and antigens for infectious disease rapid test kit
Cytokine release syndrome (CRS) is an acute systemic inflammatory syndrome characterized by fever and multiple organ dysfunction.Genemedi produces core diagnostic ingredients for test of infectious diseases and related syndrome.
GeneMedi offers paired diagnostics grade antibodies (monoclonal antibody,mab)and antigens for infectious disease rapid test kit including infection of HIV/AIDS,Hepatitis C virus(HCV), Influenza A/B,Treponema Pallidum caused Syphilis,Helicobacter pylori (H. pylori) Bacteria, Herpes simplex virus 1/2(HSV-1/HSV-2),Cytomegalovirus (CMV),rubella virus,toxoplasma gondii and so on.
All the antibodies of inflammation and CRS test are suitable for in functional ELISA, and other immunoassays in diagnostics. The antibody can act as a capture antibody and detection antibody.
Classification: Astrovirus (Aastrovirus) human adenovirus anthrax bacillus acute respiratory infectious diseases Jejunum campylobacter candida Cytomegalovirus (CMV) Cryptosporidium (Crypto) parvum EB virus ebola virus (EV) Food source pathogenic bacteria resistant FluA FluB H. pylori Hepatitis b virus (HBV) HCV HIV1 HIV2 Human papilloma virus (HPV) herpes simplex virus HSV-1 HSV-2 Listeria monocytogenes Mycobacterium tuberculosis malaria Norovirus (NV, also called Winter Vomiting Bug) Prion Rift Valley Fever (RVF) rotavirus rubella virus salmonella Sexually transmitted diseases (STDs) TP(Treponema Pallidum) toxoplasma Vaccinia virus (cowpox virus) Vibrio cholerae West Nile virus (WNV)
Reference
[1] National Institutes of Health (US) and BSC Study, Understanding Emerging and Re-emerging Infectious Diseases, National Institutes of Health (US), 2007.
[2] Nii-Trebi NI. Emerging and Neglected Infectious Diseases: Insights, Advances, and Challenges. Biomed Res Int. 2017; 2017:5245021. doi: 10.1155/2017/5245021. Epub 2017 Feb 13. PMID: 28286767; PMCID: PMC5327784.
[3] N. C. Barber and L. A. Stark, “Online resources for understanding outbreaks and infectious diseases,” CBE Life Sciences Education, vol. 14, no. 1, 2015.
[4] “Globalization and infectious diseases: a review of the linkages,” http://www.who.int/tdr/publications/documents/seb topic3.pdf.
[5] S. S. Morse, “Factors and determinants of disease emergence,” OIE Revue Scientific et Technique, vol. 23, no. 2, pp. 443–451, 2004.
[6] A. Tadin, R. Tokarz, A. Markotic et al., “Molecular survey of zoonotic agents in rodents and other small mammals in Croatia,” American Journal of Tropical Medicine and Hygiene, vol. 94, no. 2, pp. 466–473, 2016.
[7] World Health Organization, “Infections and infectious diseases: a manual for nurses and midwives in the WHO European Region,” http://www.euro.who.int/ data/assets/pdf file/0013/102316/e79822.pdf.
[8] S. S. Morse, J. A. K. Mazet, M. Woolhouse et al., “Prediction and prevention of the next pandemic zoonosis,” The Lancet, vol. 380, no. 9857, pp. 1956–1965, 2012.
[9] MATER METHODS 2013; 3:207
[10] van Elden L, Nijhuis M, Schipper P, Schuurman R, van Loon A. Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J Clin Microbiol. 2001; 39:196-200
[11] Corman V, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu D, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25
[12] Abbott. (2020b). ID NOWTM is a leading molecular point-of-care platform in the United States, trusted by hospitals, physician offices, and urgent care clinics nationwide. https://www.alere.com/en/home/product-details/id-now-covid-19.html
[13] McLoughlin K. Microarrays for pathogen detection and analysis. Brief Funct Genomics. 2011; 10:342-53
[14] Jaing C, Gardner S, McLoughlin K, Thissen J, Slezak T. Detection of adventitious viruses from biologicals using a broad-spectrum microbial detection array. PDA J Pharm Sci Technol. 2011; 65:668-74
[15] Khanna M, Kumar P, Chugh L, Prasad A, Chhabra S. Evaluation of influenza virus detection by direct enzyme immunoassay (EIA) and conventional methods in asthmatic patients. J Commun Dis. 2001; 33:163-9
[16] Waner J, Todd S, Shalaby H, Murphy P, Wall L. Comparison of Directigen FLU-A with viral isolation and direct immunofluorescence for the rapid detection and identification of influenza A virus. J Clin Microbiol. 1991; 29:479-82
[17] Lee M, Cohen B, Hand J, Nokes D. A simplified and standardized neutralization enzyme immunoassay for the quantification of measles neutralizing antibody. J Virol Methods. 1999; 78:209-17
[18] Kim M, Kang S, Lee W. Evaluation of a new rapid test kit to detect hepatitis C virus infection. J Virol Methods. 2013; 193:379-82
[19] Niu X, Wang X, Liu G, Li Y. [Establishment of the evaluation reference system for domestic anti-hepatitis C virus diagnostic enzyme immunoassay kits]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2013; 29:761-4
[20] Filice G, Soldini L, Orsolini P, Razzini E, Gulminetti R, Campisi D, et al. Sensitivity and specificity of anti-HIV ELISA employing recombinant (p24, p66, gp120) and synthetic (gp41) viral antigenic peptides. Microbiologica. 1991; 14:185-94
[21] de Boer G, Back W, Osterhaus A. An ELISA for detection of antibodies against influenza A nucleoprotein in humans and various animal species. Arch Virol. 1990; 115:47-61
[22] Cuzzubbo A, Vaughn D, Nisalak A, Solomon T, Kalayanarooj S, Aaskov J, et al. Comparison of PanBio dengue duo enzyme-linked immunosorbent assay (ELISA) and MRL dengue fever virus immunoglobulin M capture ELISA for diagnosis of dengue virus infections in Southeast Asia. Clin Diagn Lab Immunol. 1999; 6:705-12
[23] https://www.virology.ws/2010/07/16/detection-of-antigens-or-antibodies-by-elisa/
[24] Zhang WJ, Lv X, Huang C, et al. Clinical evaluation and application of detection of IgM and IgG antibodies against
SARS-CoV-2 using a colloidal gold immunochromatography assay[J]. Chinese Journal of Virology, 2020, 36(3):348-354.
[25] Li H, Li YY, Zhang ZG, et al. Establishment and clinical performance evaluation of 2019 novel coronavirus antibody colloidal gold detection method[J]. Chinese Journal of Infectious Diseases, 2020, 38(3):139-144.
[26] Augustine R, Das S, Hasan A, S A, Abdul Salam S, Augustine P, Dalvi YB, Varghese R, Primavera R, Yassine HM, Thakor AS, Kevadiya BD. Rapid Antibody-Based COVID-19 Mass Surveillance: Relevance, Challenges, and Prospects in a Pandemic and Post-Pandemic World. J Clin Med. 2020 Oct 21;9(10):3372. doi: 10.3390/jcm9103372. PMID: 33096742; PMCID: PMC7589650.
[27] LI Jia-jun, ZHENG Xiao, et al. Novel Coronavirus and Research Progress of Related Clinical Detection Methods[J]. Biotechnology Bulletin, ISSN 1002-5464, CN 11-2396/Q.
[28] Timothy M Uyeki, Henry H Bernstein, et al. Clinical Practice Guidelines by the Infectious Diseases Society of America: 2018 Update on Diagnosis, Treatment, Chemoprophylaxis, and Institutional Outbreak Management of Seasonal Influenza, Clinical Infectious Diseases, Volume 68, Issue 6, 15 March 2019, Pages e1–e47, https://doi.org/10.1093/cid/ciy866
[29] Anestad G. Surveillance of respiratory viral infections by rapid immunofluorescence diagnosis, with emphasis on virus interference. Epidemiol Infect. 1987; 99:523-31
[30] Daisy J, Lief F, Friedman H. Rapid diagnosis of influenza A infection by direct immunofluorescence of nasopharyngeal aspirates in adults. J Clin Microbiol. 1979; 9:688-92
[31] Johnson J, Higgins A, Navarro A, HUANG Y, Esper F, Barton N, et al. Subtyping influenza A virus with monoclonal antibodies and an indirect immunofluorescence assay. J Clin Microbiol. 2012; 50:396-400
[32] Fauvel M, Ozanne G. Immunofluorescence assay for human immunodeficiency virus antibody: investigation of cell fixation for virus inactivation and antigen preservation. J Clin Microbiol. 1989; 27:1810-3
[33] Klespies S, Cebula D, Kelley C, Galehouse D, Maurer C. Detection of enteroviruses from clinical specimens by spin amplification shell vial culture and monoclonal antibody assay. J Clin Microbiol. 1996; 34:1465-7
[34] Coffin S, Hodinka R. Utility of direct immunofluorescence and virus culture for detection of varicella-zoster virus in skin lesions. J Clin Microbiol. 1995; 33:2792-5
[35] Caviness A, Oelze L, Saz U, Greer J, Demmler Harrison G. Direct immunofluorescence assay compared to cell culture for the diagnosis of mucocutaneous herpes simplex virus infections in children. J Clin Virol. 2010; 49:58-60
[36] Pouletty P, Chomel J, Thouvenot D, Catalan F, Rabillon V, Kadouche J. Detection of herpes simplex virus in direct specimens by immunofluorescence assay using a monoclonal antibody. J Clin Microbiol. 1987; 25:958-9
[37] Liu I, Chen P, Yeh S, Chiang Y, Huang L, Chang M, et al. Immunofluorescence assay for detection of the nucleocapsid antigen of the severe acute respiratory syndrome (SARS)-associated coronavirus in cells derived from throat wash samples of patients with SARS. J Clin Microbiol. 2005; 43:2444-8
[38] https://fineartamerica.com/featured/6-vaccinia-virus-infected-cell-dr-dan-kalman.html
[39] Schochetman G, Epstein J, Zuck T. Serodiagnosis of infection with the AIDS virus and other human retroviruses. Annu Rev Microbiol. 1989; 43:629-59
[40] Damen M, Zaaijer H, Cuypers H, Vrielink H, van der Poel C, Reesink H, et al. Reliability of the third-generation recombinant immunoblot assay for hepatitis C virus. Transfusion. 1995; 35:745-9
[41] Kong Q, Xue C, Ren X, Zhang C, Li L, Shu D, et al. Proteomic analysis of purified coronavirus infectious bronchitis virus particles. Proteome Sci. 2010; 8:29
[42] Ren X, Xue C, Kong Q, Zhang C, Bi Y, Cao Y. Proteomic analysis of purified Newcastle disease virus particles. Proteome Sci. 2012; 10:32
[43] Nagler F, Rake G. The Use of the Electron Microscope in Diagnosis of Variola, Vaccinia, and Varicella. J Bacteriol. 1948; 55:45-51
[44] Gust I, Kaldor J, Cross G, Waugh M, Ferris A. Virus-like particles associated with a faecal antigen from hepatitis patients and with Australia antigen. Aust J Exp Biol Med Sci. 1971; 49:1-9
[45] Kapikian A, Wyatt R, Dolin R, Thornhill T, Kalica A, Chanock R. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J Virol. 1972; 10:1075-81
[46] Johnson K, Lange J, Webb P, Murphy F. Isolation and partial characterisation of a new virus causing acute haemorrhagic fever in Zaire. Lancet. 1977; 1:569-71
[47] Pattyn S, van der Groen G, Jacob W, Piot P, Courteille G. Isolation of Marburg-like virus from a case of haemorrhagic fever in Zaire. Lancet. 1977; 1:573-4
[48] Chua K, Wong E, Cropp B, Hyatt A. Role of electron microscopy in Nipah virus outbreak investigation and control. Med J Malaysia. 2007; 62:139-42
[49] Hyatt A, Zaki S, Goldsmith C, Wise T, Hengstberger S. Ultrastructure of Hendra virus and Nipah virus within cultured cells and host animals. Microbes Infect. 2001; 3:297-306
[50] Drosten C, Gunther S, Preiser W, van der Werf S, Brodt H, Becker S, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003; 348:1967-76
[51] Reed K, Melski J, Graham M, Regnery R, Sotir M, Wegner M, et al. The detection of monkeypox in humans in the Western Hemisphere. N Engl J Med. 2004; 350:342-50
[52] https://www.hantasite.com/2017/03/hantavirus-life-cycle-and-infection.html






