Investigation of novel SARS-CoV-2 variant
Variant of Concern 202012/01
Technical briefing 2
Nomenclature of variants in the UK
SARS-CoV-2 variants if considered to have concerning epidemiological, immunological or pathogenic
properties are raised for formal investigation. At this point they are designated
Variant Under Investigation (VUI) with a year, month, and number. Following risk assessment with
the relevant expert committee, they may be designated Variant of Concern (VOC). This variant was
designated VUI 202012/01 on detection and on review re-designated as VOC 202012/01 on
18/12/2020.
Current epidemiological findings
Only a small fraction of all new cases of VOC 202012/01 are identified by whole-genome
sequencing, and this data typically lags test date by approximately two weeks, therefore a proxy
S gene target failure (SGTF)) is used to indicate carriage of the VOC.
We previously observed that one of the S gene mutations in the VOC, which deletes amino acids 69
and 70 (Δ69-70), causes a reproducible SGTF in the Thermopath TaqPath assay used in three UK
lighthouse laboratories (see Technical Briefing
1). This coincidental occurrence therefore provides a good proxy for monitoring trends
in VOC 202012/01. SGTF correlates almost perfectly with presence of Δ69-70. Considering 14,950
tested samples where we know both the sequence and the SGTF status, 99.3% of Δ69-70 sequences
(1,831 of 1,843) are SGTF, compared to 0.05% of sequences without the deletion (7 of 13107).
Because Δ69-70 has arisen multiple times, and SGTF is a proxy for any lineage with that
mutation, the utility of SGTF as a proxy for VOC 202012/01 varies over time and region. Table 2
shows, for all pillar 2 sequences, the weekly proportion of Δ69-70 sequences that were confirmed
to be VOC 202012/01. Table 3 shows the proportion of Δ69-70 that is the VOC 202012/01 in England
during December, broken down by region. It is, as expected, highest in the areas where the VOC
was first observed, but it has been a substantial majority in all areas of England during the
month of December. The numbers in these tables are based on sequenced samples, some of which may
have come from the same individual (this effect is likely to be small).
| Week beginning | Per cent VOC of all Δ69-70 | Number of pillar 2 Δ69-70 sequences |
| 44116 | 0.03 | 116 |
| 44123 | 0.15 | 219 |
| 44130 | 0.29 | 156 |
| 44137 | 0.64 | 398 |
| 44144 | 0.79 | 632 |
| 44151 | 0.88 | 602 |
| 44158 | 0.93 | 372 |
| 44165 | 0.96 | 370 |
| 44172 | 0.98 | 2007 |
| Region | Per cent VOC of all Δ69-70 | Number of pillar 2 Δ69-70 sequences December 1st- 21st |
| East Midlands | 0.82 | 45 |
| East of England | 0.99 | 375 |
| London | 0.98 | 1056 |
| North East | 0.91 | 58 |
| North West | 0.93 | 83 |
| South East | 0.99 | 646 |
| South West | 1 | 34 |
| West Midlands | 0.98 | 80 |
| Yorkshire and the Humber | 0.89 | 36 |
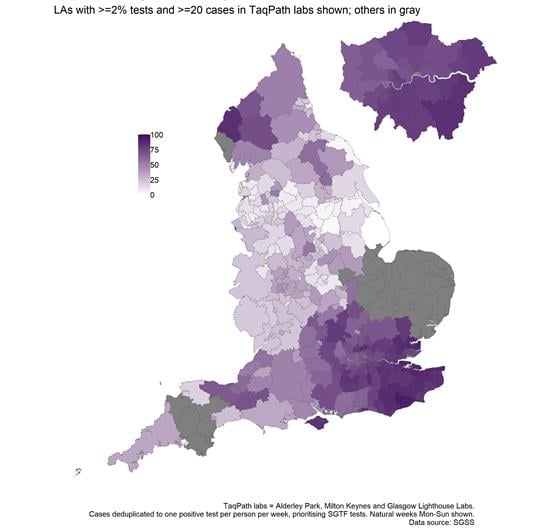
The proportion of England specimens tested in the three lighthouse laboratories using the assay
which produces the S gene drop out is substantial and relatively constant over time (Figure 1).
The
proportion of cases tested by this assay which are SGTF has continued to rise in December, in
all
age groups though most markedly in persons aged 25-49 years. The spatial distribution of SGTF
cases
shows a relatively higher burden in the South East and London, parts of the South West region
and
Cumbria.
To note, the low coverage in East of England by the three Thermofisher TaqPath labs precludes
any
analysis of SGTF burden in that region. There is less confirmatory genomic data in the North
West
and additional sequencing is being undertaken
Figure 1. Proportion of
England specimens tested in Thermofisher TaqPath, by S-gene detection by week, 1
September
to 22 December 2020
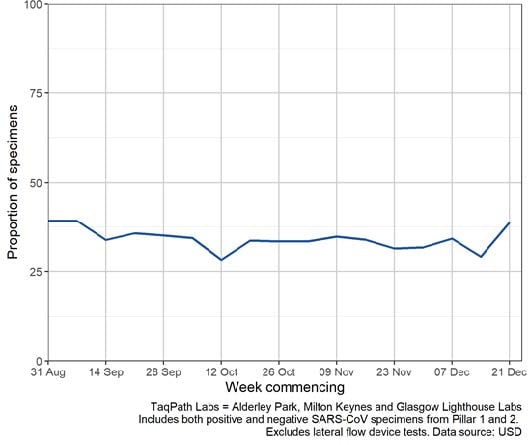
Figure 2. Weekly number
of
cases tested by laboratories using Thermofisher TaqPath, by S-gene detection
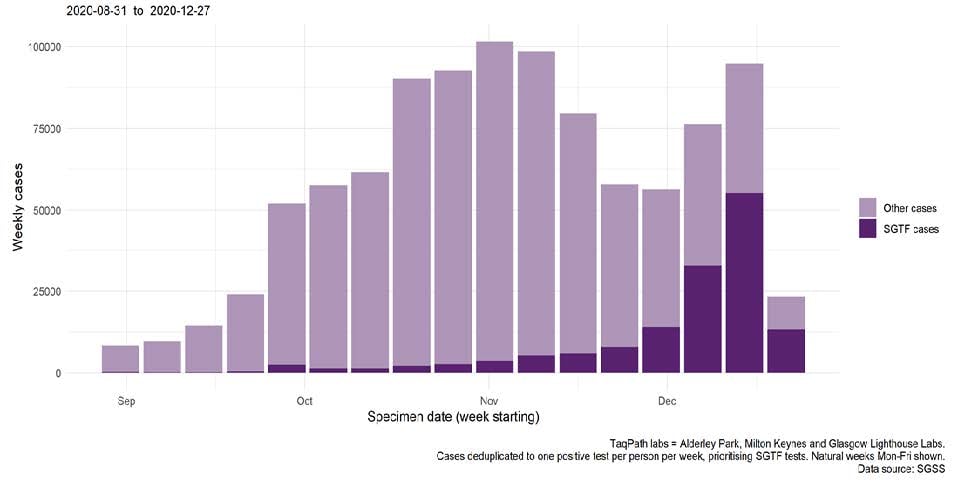
Figure 3. Weekly number
of
cases tested by laboratories using Thermofisher TaqPath, by S-gene detection and age
group
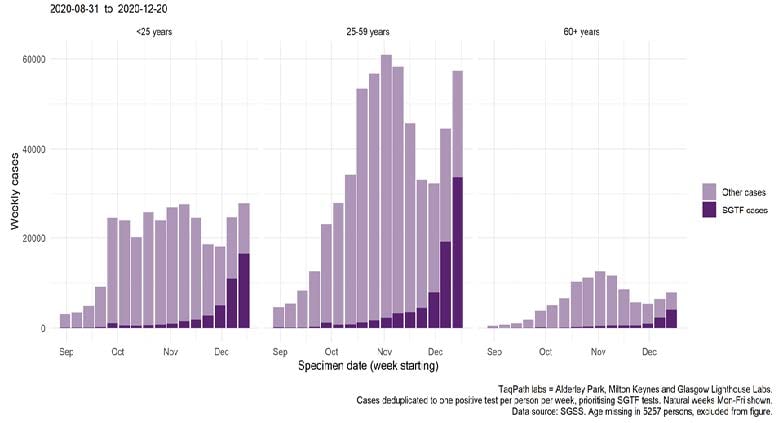
Figure 4. Age-sex
pyramid
of cases tested by laboratories using Thermofisher TaqPath, by S-gene detection
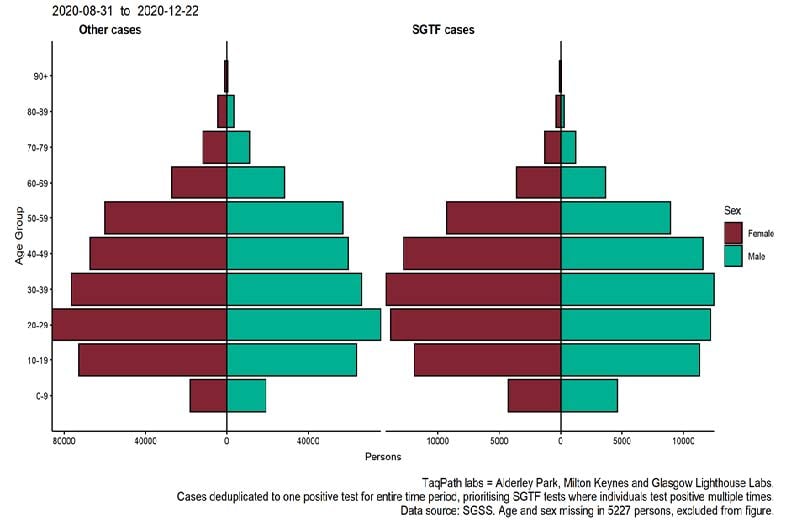
Figure 5. Proportion of
COVID-19 cases with SGTF among cases tested by laboratories using Thermofisher TaqPath,
by
S-gene detection (14 to 20 December 2020)
Preliminary findings of matched cohort study
A matched cohort study was undertaken to inform a preliminary assessment of outcomes of
hospitalisation and case fatality associated with VOC 202012/01. This analysis of sequenced
positive SARS-CoV2 cases used confirmed VOC cases matched to confirmed wild-type comparator
cases (classified as distinct to the variant sequence). To optimise comparability of the variant
(VOC 202012/01) and wild-type cases and manage the impact of non-random sampling of SARS-CoV-2
cases for sequencing, variant cases were frequency matched to wild-type cases on a 1:1 basis by
age group, sex, upper tier local authority (UTLA) of residence and two-week time-period for
specimen date.
Of the 2,693 variant cases identified at the time of analysis, 1,769 variant cases with specimen
dates between 20 September and 15 December 2020 were matched to 1,769 wild-type comparator cases
and were included in this analysis. In reflection of the matching criteria, the median age of
variant cases was 36 years and 35 for the wild-type cases. 51.4% of both the variant cases
wild-type comparator cases were female.
The comparison of the 3,538 variant and wild-type cases showed that the majority of variant
cases were of White ethnicity (75.2%) followed by Asian (10.1%) and Black (5.9%). The ethnic
profile of wild-type comparator cases was broadly similar but a higher proportion of Asian
ethnicity (13.5% v 10.1%).
The majority of variant cases were resident in private dwellings (95.0% in variant cases and
94.3% in wild-type comparator cases). Variant cases were more likely to be part of a residential
cluster (defined as all laboratory confirmed cases occurring at the same Unique Property
Reference Number (UPRN) within 14 days of each other) compared to wild-type comparator cases
(63.5% vs 56.1%, Chi-Squared test p=0.00).
Review of hospital admissions data from the NHS identified that of the 3,538 cases, 42
individuals had a record of hospital admission after the date of specimen. Fewer variant cases
(16 cases (0.9%)) were admitted to hospital compared to wild-type comparator cases (26 cases
(1.5%)) but the difference was not significant (Chi-squared test p=0.162). Due to potential time
delays for receipt of hospital admissions data, the identified hospital admissions should be
regarded as a minimum number of hospital admissions and further admissions data are likely to be
received into this NHS dataset in the future.
The 28-day case fatality was assessed for variant cases and comparator cases.
Analysis was restricted to 2,700 cases with a full 28 days elapsed since the specimen date.
Among variant cases, 12 of 1,340 (0.89%) variant cases died within 28 days of their specimen
date compared with 10 of 1,360 (0.73%) wild-type comparator cases; this difference was not
signfiicant (Odds ratio:1.21, p=0.65).
Re-infection
Laboratory data were used to identify possible reinfections; these were defined as an episode of
polymerase chain reaction (PCR) positivity at least 90 days before a recent PCR positive
detection. Two reinfections were detected in in the variant case group (1.13/1000 cases)
compared to 3 reinfections in the comparator group (1.70/1000 cases, Fisher’s exact P=1.00).
The same definition was applied to SGTF cases. These SGTF cases included samples with orf and N
gene Ct values of <31, S gene defined as negative or positive based on presence or absence
regardless of Ct value. The rate of detected re-infections in national SGTF cases was
0.60/1000 compared to a rate of 0.61/1000 in non-SGTF cases (P=0.94). When limited
geographically to the Kent area, the rate of detected re-infections was 0.51/1000 compared
to 0/1000 in non-SGTF cases (P=0.69).
Secondary attack rates
We investigated secondary attack rates using data from NHS Test and Trace, the national
contact tracing system in England. Between 5 October and 6 December 2020, 1,105,388
cases were reported to NHS Test and Trace; 46,237 (4.2%) had genomic sequencing data
(around 700 (1-2%) poor quality). 1,978 had the variant (VOC 202012/01), 4.3% of those
with sequencing data.
228,361 (9.9% attack rate) of all contacts notified by cases in this period became
cases:
• 15.1% among those whose index case was confirmed to have the VOC 202012/01
• 9.8% among those whose index case was sequenced and confirmed with other variants
Summary
The findings from the analysis of data on SGTF cases showed a roughly similar spatial
distribution of cases as observed from mapping of genomic data. SGTF cases were mostly
observed in the South East, London parts of the South West regions and Cumbria. This
regional variation in should be interpreted in the context of limited coverage in
regions like East of England of by the three Thermofisher TaqPath lighthouse labs.
However, in regions with relatively high and consistent coverage, the findings can be
used as a proxy for the burden of VOC 201212/01 infection.
Preliminary results from the cohort study found no statistically significant difference
in hospitalisation and 28-day case fatality between cases with the variant (VOC
201212/01) and wild-type comparator cases. There was also no significant difference in
the likelihood of reinfection between variant cases and the comparator group.
Data sources
Data used in this investigation is derived from the COG-UK dataset, the PHE Second Generation Surveillance System, the secondary uses service (SUS) dataset and Emergency Care Data Set (ECDS).
GISAID reference genome
Sequences from this VOC can be identified by searching for the B1.1.7 lineage on GISAID (gisaid.org). The canonical VOC genome is deposited with accession EPI_ISL_601443.






