AAV-LC3 production service for Autophagy Flux Detection
Order Information
-
Step 1:
AAV vector construction -
Step 2:
AAV packaging -
Step 3:
Custom Order confirmation
| Buy | Catalog No. | Product Name | Lead time(workday) |
| GM-AAVPAU01 | AAV-CMV-mRFP-GFP-LC3 | 3 | |
| GM-AAVPAU02 | AAV-CMV-GFP-LC3 | 3 |
| Buy | AAV Vector Production Scale | Final Deliverables | Lead Time(workday) |
|---|
Introduction to AAV-LC3 production service for Autophagy Flux Detection
Premade LC3 Autophagy Biosensors Products and user manual
Adeno associated virus(AAV) AAV-GFP-LC3 Autophagy BiosensorAAV-mRFP-GFP-LC3 Autophagy Biosensor |
Adenovirus Adv-GFP-LC3 Autophagy BiosensorAdv-mRFP-GFP-LC3 Autophagy Biosensor |
Lentivirus Lv-GFP-LC3 Autophagy BiosensorLv-mRFP-GFP-LC3 Autophagy Biosensor |
LC3 Autophagy Biosensors User Manual
AAV-LC3 User Manual  |
Adenovirus-LC3 User Manual  |
Lentivirus-LC3 User Manual  |
Autophagy
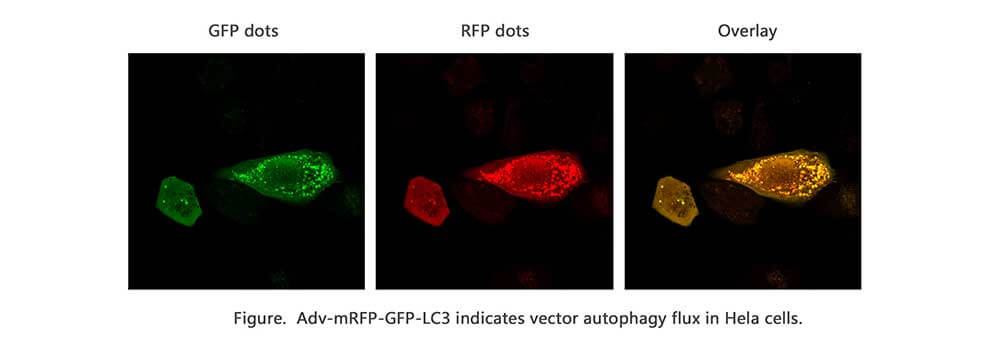
Autophagy is a self-degradative process in cell that is important for balancing sources of energy at critical times in development and in response to nutrient stress. For autophagy study, Genemedi supply autophagy biosensor, in which GFP and/or RFP tags are fused at the C-termini of the autophagosome marker LC3, allowing to detect the intensity of autophagy flux in real-time with more accuracy, clarity and intuitiveness. These biosensors provide an enhanced dissection of the maturation of the autophagosome to the autolysosome, which capitalizes on the pH difference between the acidic autolysosome and the neutral autophagosome. The acid-sensitive GFP will be degraded in autolysosome whereas the acid-insensitive RFP will not. Therefore, the change from autophagosome to autolysosome can be visualized by imaging the specific loss of the GFP fluorescence, leaving only red fluorescence. Adeno associated virus (AAV)
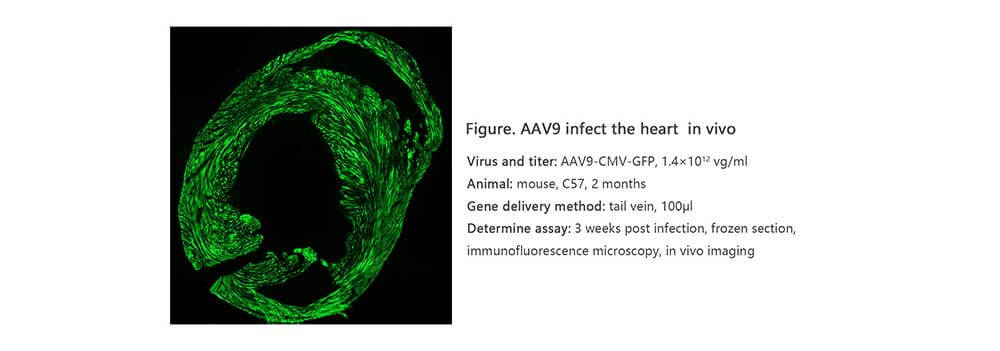
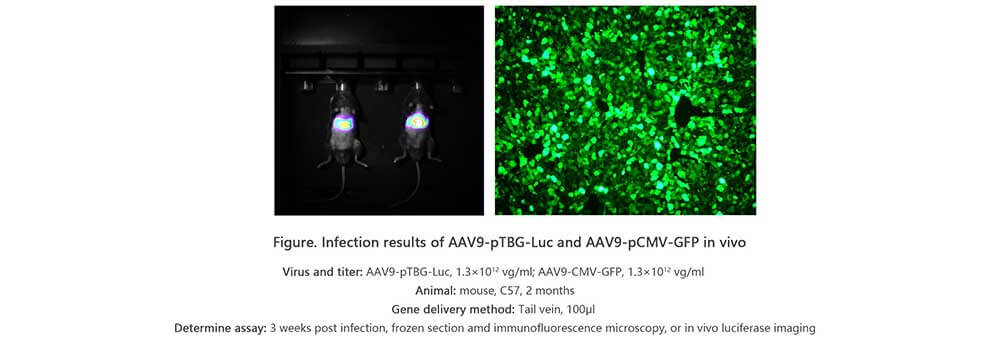
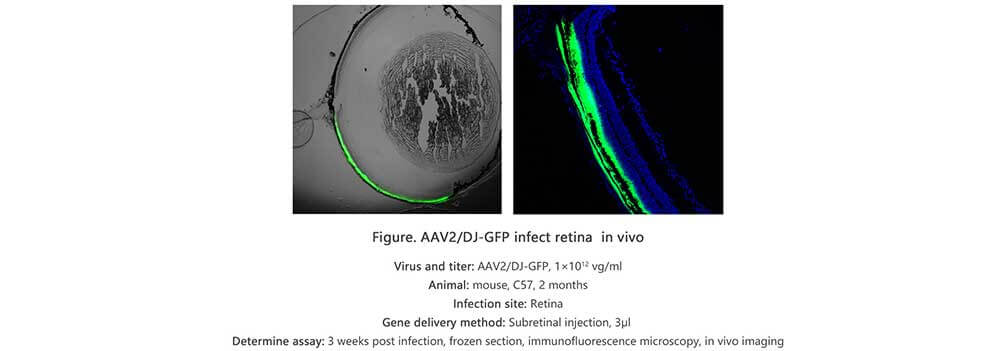
Adeno associated virus (AAV) is one kind of human parvovirus. Recombinant AAV vectors can efficiently transfect various cell types, including dividing and quiescent cells, and induce persistent gene expression in vivo without integrating into host genome and causing any disease. These features make AAV an attractive candidate in the application of gene delivery for gene therapy and human disease model establishment. To date, AAV has been proved as the most excellent gene therapy vector. Over 204 clinical trials have been carried out using AAV vectors for gene delivery, and promising gene therapy outcomes have been achieved from clinical trials for a great number of diseases. AAV-LC3 production service for Autophagy Flux Detection
Taking advantage of RFP-GFP-LC3 and GFP-LC3 labeling system, Genemedi has launched the production service of AAV-RFP-GFP-LC3 and AAV-GFP-LC3, which can be used to observe autophagy flux and monitor the intensity of autophagy flux in real-time in vivo or in vitro.
Properties
| AAV-RFP-GFP-LC3 OR AAV-GFP-LC3 | |
|---|---|
| Quantity/Unit | Vials |
| Form | Frozen form |
| Suitable Types of Infection | In vivo infection in animals |
| Sipping and Storage Guidelines | Shipped by dry ice, stored at -80 ℃, effective for 1 year. Avoid repeatedly freezing and thawing. |
| Titer | > 1*10^12v.g/ml |
Advantages
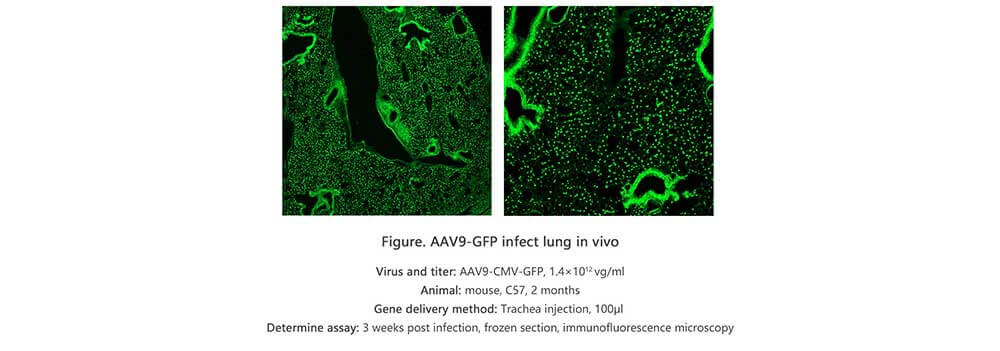
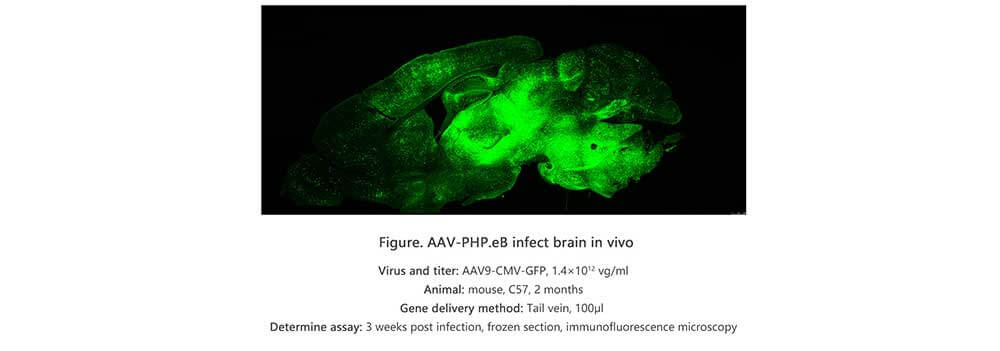
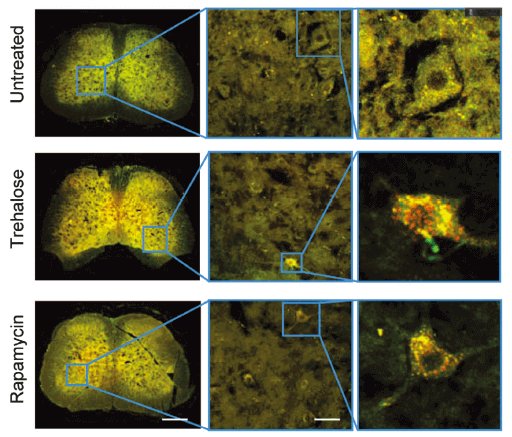
Applications and Figures
Quality control description
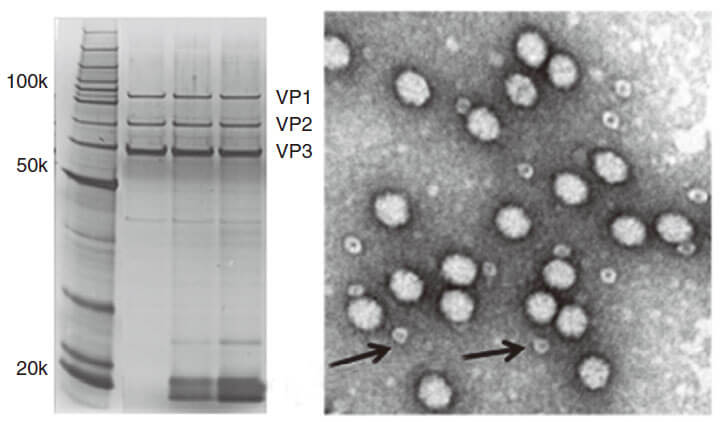 Figure 3. PAGE for quality testing example of AAV by Genemedi. Only 3 protein bands can be seen in the purified AAV virus samples.
Figure 3. PAGE for quality testing example of AAV by Genemedi. Only 3 protein bands can be seen in the purified AAV virus samples.Technical Documents
 AAV(adeno associated virus) User Manual.
2. For more information about how to use in vivo Autophagy Flux Detection with AAV vectors, please refer to the
AAV(adeno associated virus) User Manual.
2. For more information about how to use in vivo Autophagy Flux Detection with AAV vectors, please refer to the  AAV-LC3 Autophagy Flux Detection Manual.
AAV-LC3 Autophagy Flux Detection Manual.
Frequently Asked Questions(FAQs)
- 1. How can I choose the optimal AAV serotypes for my in vivo study?
-
AnswerGenemedi has launched a comprehensive AAV production service. More than 12 AAV serotypes and a variety of capsid engineered AAV vectors are available for targeting different tissues and organs. Different AAV serotypes have different tissue tropism in vivo, the common serotypes and their tropism are listed in the above table 1. Therefore, you can select the most suitable AAV serotype for your study according to this table. For example, if you’d like to transfect your target gene into mouse ears, the AAV-Anc80 will be your best choice. For your optimal AAV serotypes screening, Genemedi can also customize an AAV serotype testing kit with recommended serotypes of your choice.
- 2. How much DNA do I need to provide for Custom AAV without DNA amplification?
-
AnswerYou will need to provide purified plasmid DNA at a concentration of 0.5ug/ul or more. 50 ug DNA needed for Custom AAV production service without DNA amplification (10^9 GC/ml). 200 ug DNA needed for Custom AAV production service without DNA amplification (10^12 GC/ml). 300 ug DNA needed for Custom AAV production service without DNA amplification (10^13 GC/ml).
- 3. What's the difference of AAV titer unit between GC/ml and vg/ml?
-
AnswerThe AAV titer unit GC/ml and vg/ml can be used interchangeably, based on qPCR method.
- 4. Why can’t I see the expression of GFP after AAV infection in my cell line?
-
AnswerAAV serotypes selection is an important parameter which may affect the transduction ability of AAV particles. Thus, it is necessary to determine which serotype works best for your cell line. For instance, serotype 5 limits its transduction ability on most cell types. For detailed information, you can refer to our technical sheet "AAV - General Guideline to Serotype Selection".
- 5. Does Genemedi’s AAV preparations belong to in-vivo grade?
-
AnswerExtensive purification steps guarantee the high quality of our viral particles, which are ready to be administrated for in-vivo models. For detailed references, please see the in-vivo infection data tested by another independent lab { www.genemedi.net }
- 6. What is the QC (quality control) method for testing AAV in Genemedi?
-
AnswerWe provide qPCR-based titer as the primary method to determine whether the packaging/purification process is successful or not. If your virus has GFP or RFP reporter, we also perform virus infectivity testing in HEK293T cell line.
- 7. Can I utilize different serotypes of AAV virus in the same equipment to keep the infected cells?
-
AnswerThere is no problem with using different serotypes in the same equipment, as long as the handler takes the basic precautions to avoid cross-contamination.
- 8. What is the difference between brain localization of gene expression after injection of AAV or lentiviral vectors?
-
AnswerLentiviral particles don't spread well after stereotaxic injection into brain because the particles are relatively large (90nm~100nm). In contrast, AAV particles spread more readily due to smaller size (20nm~100nm). As mentioned, it has been stated that some AAV serotypes spread better than others in brain, for example, AAV5 is reported to spread exceptionally well when injected into striatum. Lastly, pretreatment with mannitol to your animal about 15 min ahead of viral injection has been reported to aid in spread of viral particles in the brain.
- 9. Do AAVs tranduce axons passing through a region of interest?
-
AnswerAAVs are able to infect axon terminals and produce retrograde transport (towards the cell body). The slightly longer answer is that this process is highly biased based on the serotype you are using. There are a number of papers reporting AAV6 (and maybe AAV6.2) is the main serotype to produce retrograde transport. There are however reports of many other serotypes producing retrograde transport with a much smaller rate.
- 10. How can I increase the transduction efficiency?
-
AnswerTry varying particle-to-cell ratio (PPC), incubation volume, temperature and, cell density (if adherent cells are transduced). For adherent cells, we recommend a confluence of about 70%. Following the PPC, adjusting the volume is the next best parameter to change to optimize protein expression.
- 11. What is the difference between LC3A, LC3B and LC3C, or LC3-I and LC3-II?
-
AnswerLC3 is a soluble protein with a molecular mass of ∼17 kD and is distributed ubiquitously in eukaryotes. It is expressed as the splice variants LC3A, LC3B, and LC3C which display unique tissue distribution. All LC3 isoforms undergo post-translational modifications, especially PE conjugation (lipidation) during autophagy. Upon autophagic signal, the cytosolic form of LC3 (LC3-I) is conjugated to PE to form LC3-PE conjugate (LC3-II), which is recruited to the autophagosome membranes.
- 12. Will the GFP signal of mRFP-GFP-LC3 that anchored at autophagosomes and fused with lysosome relight after the cell fixed?
-
AnswerGFP is no longer delighted reversibly once GFP-LC3 signal was destroyed by the acidic environment.
- 13. Can transient expression of RFP-GFP-LC3 differ badly from stable expression?
-
AnswerTransient transfection of GFP-LC3 leads to the formation of non-autophagic LC3 puncta due to overexpression. Reports shows that GFP-LC3 is aggregate prone protein. We suggest the use of stable cell line for assessment of GFP-LC3 puncta.
- 14. Which software do you usually use for counting the number of LC3 dots in the cell?
-
AnswerImage J does work. For LC3 puncta, we suggest using the "Watershed" plugin from Image J. But proper threshold adjustment is critical. CellProfiler is another choice. First use the top hat filter. Then with the Identify Primary Objects module, manually adjust the Intensity threshold to optimally select the punctas of the right intensity.
- 15. How long is half-life time of chloroquine when I treat a cell line for autophagy study?
-
AnswerChloroquine is an attractive drug agent effective for the treatment of not only malaria but also inhibition of autophagy, which is a promising effect for anti-tumor therapy. Half-life time of this drug is approximately 18 hours in vivo due to the degradation by the liver, but the stability of chloroquine in vitro experiments is expected to be longer. It will depend mainly on the cell line you are working on and their metabolic activity. For example, HepG2 cells (human hepatocarcinoma cells) have a strong capacity to metabolize drugs, which might not be the case for other cell line of different tissular origin. I would suggest to add chloroquine when you change your growth medium.
- 16. Which medium is best to use to induce LC3 puncta in HeLa cells?
-
AnswerWe recommend the media used by Axe et al. (2008) JCB 182: 685-701. The recipe is 140 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 5 mM glucose, and 20 mM Hepes, pH 7.4, Add 1% BSA and pass through a 20um filter before use.
- 17. What is the best applicable inhibitor of autophagy?
-
AnswerI prefer to use CQ (chloroquine) in all of my researches about autophagy inhibition. The using of CQ is quite easy, since it is easy dissolve in water (unlike 3-MA in DMSO). One time I have been used 3-MA, actually I did not like to work with it. Bafilomycin is another choice for you.
- 18. When to add bafilomycin to study autophagy?
-
AnswerIt is better to do a time course to make sure. We use at least two times (2 and 4 hours) to starve the cells incubate bafilomycin for 2 hours (for 4 hours: the 2 last hours). Then LC3-II and P62 are detected by WB. For the degradation of p62, sometimes, 4 hours is better than 2 hours.
- 19. Which neuronal cell line should I use for autophagy level?
-
AnswerSHSY5Y and PC12 are two well established neuronal cell lines, human and rat cell lines, respectively. You may also try Neuro2A, it is a mouse cell line.
- 20. How to distinguish selective and non-selective autophagy?
-
AnswerYou can analyze selective mitophagy by interaction/co-localization study of mitochondrial marker protein and the autophagy specific adaptor protein LC3II.
- 21. When to add the autophagy inducers or inhibitors in cell culture models?
-
AnswerFirst, some cells are very sensitive to these inhibitors and you need to optimize your conditions. The second point, if you like to stop autophagy process after inducing it to compare whether it is a cell death or cell survival you may add it just hours (2-3) before treatment. However, many articles prefer to use it as pre-treatment 1 hour before any other treatment.
Reference
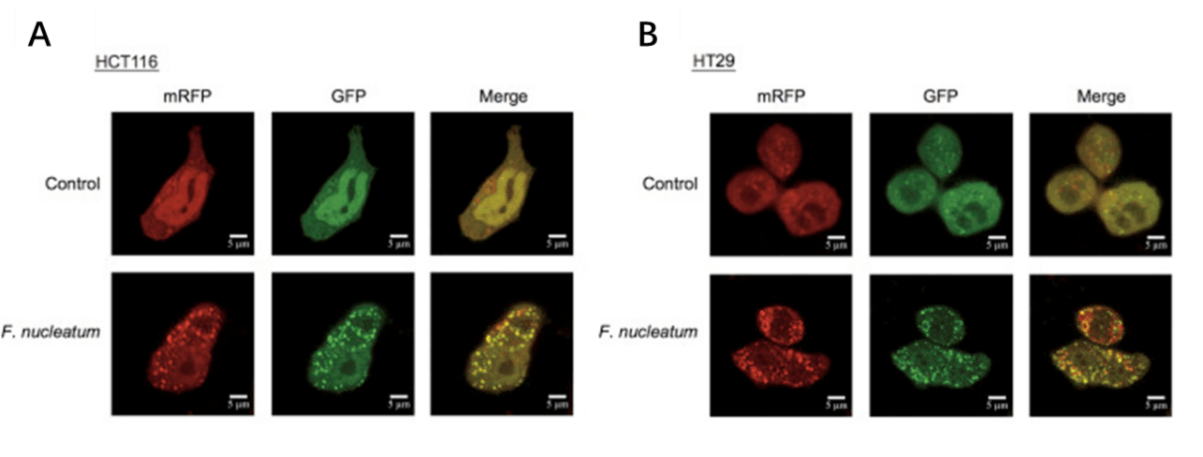
1. Yu T, F Guo, Y Yu, T Sun, D Ma, J Han, Y Qian, I Kryczek, D Sun, N Nagarsheth, Y Chen, H Chen, J Hong, W Zou and JY Fang. (2017). Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 170:548-563 e16.
2. Yuan Y, Y Zheng, X Zhang, Y Chen, X Wu, J Wu, Z Shen, L Jiang, L Wang, W Yang, J Luo, Z Qin, W Hu and Z Chen. (2017). BNIP3L/NIX-mediated mitophagy protects against ischemic brain injury independent of PARK2. Autophagy 13:1754-1766.
3. Shao BZ, P Ke, ZQ Xu, W Wei, MH Cheng, BZ Han, XW Chen, DF Su and C Liu. (2017). Autophagy Plays an Important Role in Anti-inflammatory Mechanisms Stimulated by Alpha7 Nicotinic Acetylcholine Receptor. Front Immunol 8:553.
4. Liu M, K Liang, J Zhen, M Zhou, X Wang, Z Wang, X Wei, Y Zhang, Y Sun, Z Zhou, H Su, C Zhang, N Li, C Gao, J Peng and F Yi. (2017). Sirt6 deficiency exacerbates podocyte injury and proteinuria through targeting Notch signaling. Nat Commun 8:413.
5. Chen ZH, WT Wang, W Huang, K Fang, YM Sun, SR Liu, XQ Luo and YQ Chen. (2017). The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ 24:212-224.
6. Zhang Y, K Shen, Y Bai, X Lv, R Huang, W Zhang, J Chao, LK Nguyen, J Hua, G Gan, G Hu and H Yao. (2016). Mir143-BBC3 cascade reduces microglial survival via interplay between apoptosis and autophagy: Implications for methamphetamine-mediated neurotoxicity. Autophagy 12:1538-59.
7. Luo T, J Fu, A Xu, B Su, Y Ren, N Li, J Zhu, X Zhao, R Dai, J Cao, B Wang, W Qin, J Jiang, J Li, M Wu, G Feng, Y Chen and H Wang. (2016). PSMD10/gankyrin induces autophagy to promote tumor progression through cytoplasmic interaction with ATG7 and nuclear transactivation of ATG7 expression. Autophagy 12:1355-71.
8. Hua F, K Li, JJ Yu, XX Lv, J Yan, XW Zhang, W Sun, H Lin, S Shang, F Wang, B Cui, R Mu, B Huang, JD Jiang and ZW Hu. (2015). TRB3 links insulin/IGF to tumour promotion by interacting with p62 and impeding autophagic/proteasomal degradations. Nat Commun 6:7951.
9. Zhang X, Y Yuan, L Jiang, J Zhang, J Gao, Z Shen, Y Zheng, T Deng, H Yan, W Li, WW Hou, J Lu, Y Shen, H Dai, WW Hu, Z Zhang and Z Chen. (2014). Endoplasmic reticulum stress induced by tunicamycin and thapsigargin protects against transient ischemic brain injury: Involvement of PARK2-dependent mitophagy. Autophagy 10:1801-13.
10. Li C, W Sun, C Gu, Z Yang, N Quan, J Yang, Z Shi, L Yu and H Ma. (2018). Targeting ALDH2 for Therapeutic Interventions in Chronic Pain-Related Myocardial Ischemic Susceptibility. Theranostics 8:1027-1041.
11. Yuan Y, Y Zheng, X Zhang, Y Chen, X Wu, J Wu, Z Shen, L Jiang, L Wang, W Yang, J Luo, Z Qin, W Hu and Z Chen. (2017). BNIP3L/NIX-mediated mitophagy protects against ischemic brain injury independent of PARK2. Autophagy 13:1754-1766.
12. Li S, X Dou, H Ning, Q Song, W Wei, X Zhang, C Shen, J Li, C Sun and Z Song. (2017). Sirtuin 3 acts as a negative regulator of autophagy dictating hepatocyte susceptibility to lipotoxicity. Hepatology 66:936-952.
13. Li L, B Li, M Li, C Niu, G Wang, T Li, E Krol, W Jin and JR Speakman. (2017). Brown adipocytes can display a mammary basal myoepithelial cell phenotype in vivo. Mol Metab 6:1198-1211.
14. Feng D, B Wang, L Wang, N Abraham, K Tao, L Huang, W Shi, Y Dong and Y Qu. (2017). Pre-ischemia melatonin treatment alleviated acute neuronal injury after ischemic stroke by inhibiting endoplasmic reticulum stress-dependent autophagy via PERK and IRE1 signalings. J Pineal Res 62.
15. Du X, H Hao, Y Yang, S Huang, C Wang, S Gigout, R Ramli, X Li, E Jaworska, I Edwards, J Deuchars, Y Yanagawa, J Qi, B Guan, DB Jaffe, H Zhang and N Gamper. (2017). Local GABAergic signaling within sensory ganglia controls peripheral nociceptive transmission. J Clin Invest 127:1741-1756.
16. Yang H, J Yang, W Xi, S Hao, B Luo, X He, L Zhu, H Lou, YQ Yu, F Xu, S Duan and H Wang. (2016). Laterodorsal tegmentum interneuron subtypes oppositely regulate olfactory cue-induced innate fear. Nat Neurosci 19:283-9.
17. Wu X, X Wu, Y Ma, F Shao, Y Tan, T Tan, L Gu, Y Zhou, B Sun, Y Sun, X Wu and Q Xu. (2016). CUG-binding protein 1 regulates HSC activation and liver fibrogenesis. Nat Commun 7:13498.
18. Wei Y, Y Chen, Y Qiu, H Zhao, G Liu, Y Zhang, Q Meng, G Wu, Y Chen, X Cai, H Wang, H Ying, B Zhou, M Liu, D Li and Q Ding. (2016). Prevention of Muscle Wasting by CRISPR/Cas9-mediated Disruption of Myostatin In vivo. Mol Ther 24:1889-1891.
19. Zhang X, Y Yuan, L Jiang, J Zhang, J Gao, Z Shen, Y Zheng, T Deng, H Yan, W Li, WW Hou, J Lu, Y Shen, H Dai, WW Hu, Z Zhang and Z Chen. (2014). Endoplasmic reticulum stress induced by tunicamycin and thapsigargin protects against transient ischemic brain injury: Involvement of PARK2-dependent mitophagy. Autophagy 10:1801-13.