An Insight of comparison between COVID-19 (2019-nCoV) and SARS-CoV in pathology and pathogenesis
 Full-text Download
Full-text Download
Author:
Xiaolong Cai
1. Hanbio Research Center, Hanbio Tech Co. Ltd., Shanghai, P.R. China (www.hanbio.net)
2. R&D Center, GeneMedi Co.Ltd., Shanghai, P.R. China (www.genemedi.net)
COVID-19, a novel pathogenic coronavirus emerged in China and spread globally rapidly. COVID-19 shares above 85% identity in genome with SARS-CoV. Patients infected by COVID-19 and SARS-CoV also reveal similar clinical characteristics. Here we compare the clinical and pathological features between patients infected by COVID-19 and SARS-CoV respectively.
Conclusions:
1. Older patients refer to higher case fatality rate (CFR) than young.
2. Males show a higher CFR than females, and this difference may converge as age increase.
3. COVID-19 infection may have a kidney and testis damage. Combined with higher CFR in males, genitourinary system disorder caused by the COVID-19 infection need to be cautioned.
4. It is critical to control the cytokine release syndrome(CRS) in NCP. IL-6, IL-10 and their receptors may be the drugable target.
5. Consistently to decrease of CD4+T and CD8+T cells, spleen damage, and lymphocyte depletion may exist in NCP patients. Approaches for T cell rescued may be considered.
6. Compared with SARS-CoV’s Spike protein, COVID-19 Spike protein present a higher binding affinity to ACE2, which suggests that soluble ACE2 might be a potential candidate for COVID-19 treatment. Other receptors, such as L-SIGN and DC-SIGN, need to be investigated in the future.
Introduction
A novel coronavirus pneumonia (NCP) caused by a novel coronavirus, 2019-nCoV, which named COVID-19 by WHO recently, emerged in Wuhan, Hubei province, China in December 2019. The NCP then broke-out aggressively in Jan 2020 following the human flow from Wuhan to other cities during the vocation of the Chinese Spring Festival.
Several coronaviruses can cause light respiratory disease in humans [1]. In contrast, the SARS-CoV, which emerged in 2003 in Guangdong Province, China, and MERS-CoV, in 2016, both proved host by bat and transmitted from other animals to humans, can cause severe respiratory diseases respectively[2].
Genetics Similarity of COVID-19 and SARS-CoV
The COVID-19 is also a novel coronavirus transmitted from uncertain wild animals, that can cause acute respiratory disease (ARD) with complicated clinical characteristics[3-7]. According to the whole genome sequence analysis, the COVID-19 is closer to the SARS-like bat CoVs (MG772933) than the SARS-CoV[8], which is descended from SARS-like bat CoVs.
However, COVID-19 share above 85% identity with SARS-CoV[8], but less related to MERS-CoV[9]. Importantly, within high similarity of RBD in Spike-protein, several analyses reveal that COVID-19 uses the same receptor of SARS-CoV - the angiotensin-converting enzyme 2 (ACE2)[10-12]. Meanwhile, DPP4 (dipeptidyl peptidase 4, also known as CD26), the MERS-CoV’s primary receptor, is proved not to be a receptor of COVID-19[11].
Case fatality rate (CFR) of COVID-19 and SARS-CoV both affected by age and gender.
Age and gender distinction affect the CFR of NCP and SARS respectively(Table1). It is certain that older patients refer to higher CFR than young both in NCP[3] and SARS[13, 14] respectively. Consistently to SARS, Males seem to have a higher CFR than females in NCP3, 13, 14]. Meanwhile, a SARS related study in Hongkong indicates a gradual decrease of gender-depended difference in CFP as the age increase, which may also be likely to exist in NCP[14].
Comparison of Major Pathological Characteristics between COVID-19 and SARS-CoV
We summarize the major Major Pathological Characteristics between COVID-19 and SARS-CoV in Table2. Since lots of mentions in other articles, NCP induced acute respiratory distress syndrome(ARDS) will not be discussed in this article.
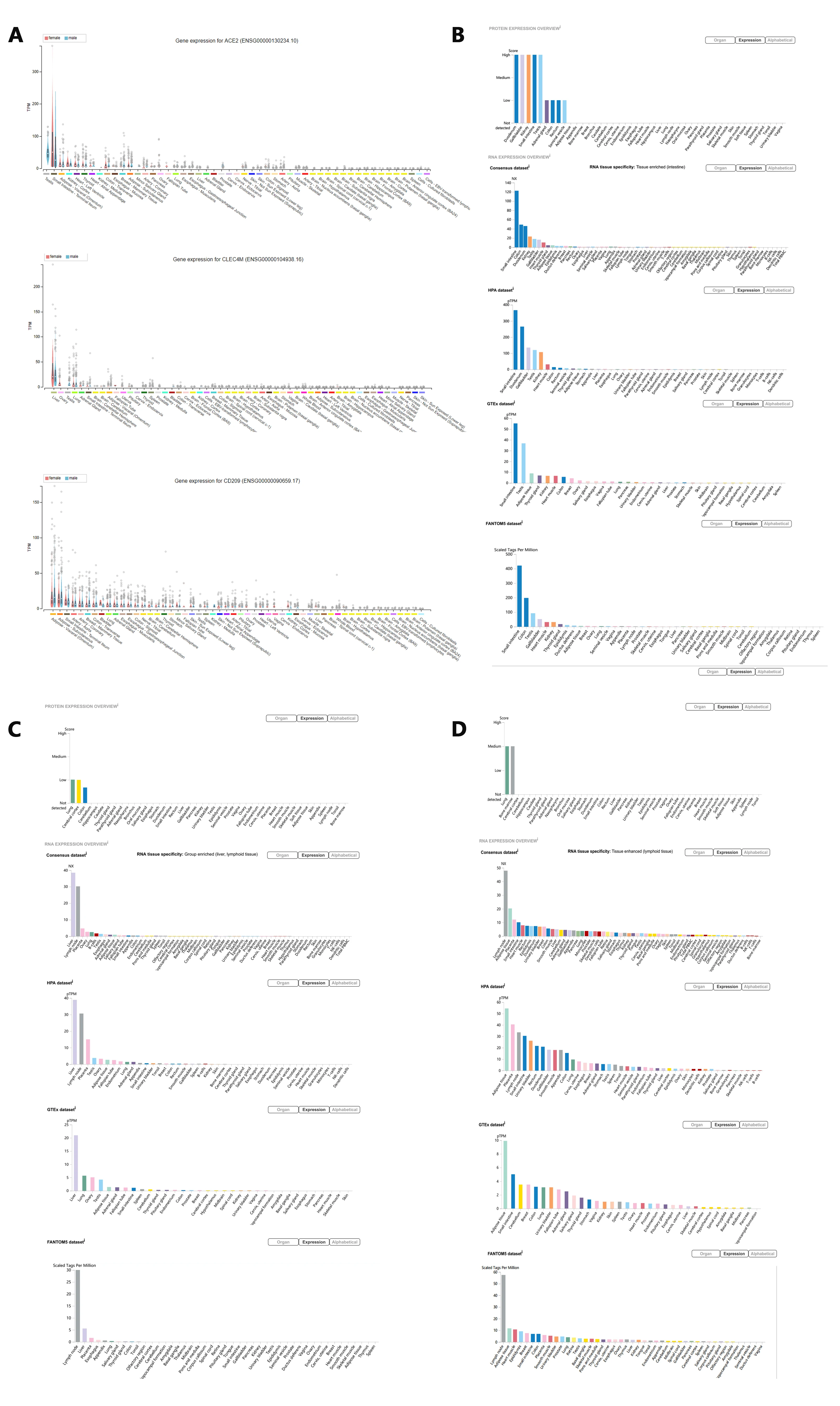
The high rate of renal impairment was observed in NCP patients, indicating the development of kidney dysfunction[15], which was consistent with SARS patients[16-20]. As is shown in Figure1, ACE2, the co-receptor of COVID-19 and SARS-CoV, presents a high expression level in the gastrointestinal tract, kidney, and testis. Since SARS induce severe testis damage[21], it holds a high risk that COVID-19 infection may also lead to testicular lesions in males,which need further clinical investigation. Considering the higher CFR of NCP and SARS in younger males than females, with the high ratio of kidney damage[12, 15](worthy of future investigation in gender), we should pay attention to the genitourinary system disorder caused by the COVID-19 infection.
Cytokine release syndrome(CRS) remains a core factor that aggravates disease progression[22]. A higher value of IL-6 and IL-10 was observed in NCP patients parallel with the severity of the disease[22]. IL-6 and IL-10 are the core cytokines that are consistently found to be elevated in patients with CRS(23,24). Tocilizumab, a monoclonal antibody targeting the IL-6 receptor, was already registered for a Clinical Trials in Anhui Provincial Hospital now (Chinese Clinical Trials Registration number: ChiCTR2000029765).
Necrosis of the spleen and atrophy of the white pulp with severe lymphocyte depletion have been mentioned in SARS patients[16-18, 25, 26]. Consistently, the amount of various immune cells including CD4+T, CD8+T, dendritic cells(DCs), macrophages, and natural killer cells(NKCs) decrease respectively[26]. Since a significant decrease of CD4+T and CD8+T cells number were also found in NCP patients respectively[22], the spleen impairment needs to be further confirmed. Meanwhile, it was reported that CD4+T cells, but not CD8+T cells are Important in control of SARS-CoV infection[27]. We might further consider different approaches for T cell rescuing within the control of CRS.
Receptors
ACE2 is proved a co-Receptor of COVID-19 and SARS-CoV. Structure analysis of COVID-19 and SARS-CoV recently indicated that COVID-19 Sike protein(S-protein) binds ACE2 with above 10 folds higher affinity than SARS-CoV[28]. This discovery further explains the more rapid transmission characteristic of the COVID-19 in humans than SARS-CoV. Referring to the higher affinity of COVID-19 S-protein and ACE2, soluble ACE2 might be a potential candidate for COVID-19 treatment.
Recognized ACE2 as the receptor of COVID-19, now scientists find important an ACE2 downstream target-TMPRSS, which maybe drugable[11, 29]. Besides ACE2, the L-SIGN (CD209L, gene symbol: CLEC4M) and DC-SIGN (CD209, gene symbol: CD209) have been identified as alternative SARS-CoV receptors respectively [30, 31]. The organ-specific expression of L-SIGN and DC-SIGN is present in Figure1C, D respectively. It is unknown if L-SIGN and DC-SIGN can act as alternative receptors of COVID-19. Regarding 14 AA sequences of RBD of SARS-CoV , COVID-19 reveals 8 strictly conserved residues and 6 AA mutations, which may affect the tropism and transmission property. Excluding ACE2, other receptors may also be available.
Concluding remarks of COVID-19 compared with SARS
1. Older patients refer to higher CFR than young.
2. Males show a higher CFP than females, and this difference may converge as age increase.
3.COVID-19 infection may have a kidney and testis damage. Combined with higher CFR in males, genitourinary system disorder caused by the COVID-19 infection need to be cautioned.
4. It is critical to control the CRS in NCP. IL-6, IL-10 and their receptors may be the drugable target.
5.Consistently to decrease of CD4+T and CD8+T cells, spleen damage, and lymphocyte depletion may exist in NCP patients. Approaches for T cell rescued may be considered.
6. Other receptors, such as L-SIGN and DC-SIGN, need to be investigated in the future.
Acknowledgement
Tribute to the medical staff fighting against the COVID-19 in China. You are the angels.
Thanks for the helps from my colleagues from Hanbio Tech Co. Ltd., Shanghai, P.R. China (www.hanbio.net) and GeneMedi Co.Ltd., Shanghai, P.R. China (www.genemedi.net) respectively.
Reference
1.
L.E.a.V.D.M. Gralinski, Return of the Coronavirus: 2019-nCoV. , Viruses, 2020. 12(2). (2020).
2.
J.L. V. M. Corman, M. Witzenrath, Coronaviruses as the cause of respiratory infections, Internist (Berl) 60, 1136-1145 (2019).
3.
Y. Yang, Q. Lu, M. Liu, Y. Wang, A. Zhang, N. Jalali, N. Dean, I. Longini, M.E. Halloran, B. Xu, X. Zhang, L. Wang, W. Liu, L. Fang, Epidemiological and clinical features of the 2019 novel coronavirus outbreak in China, medRxiv, (2020).
4.
J. Li, S. Li, Y. Cai, Q. Liu, X. Li, Z. Zeng, Y. Chu, F. Zhu, F. Zeng, Epidemiological and Clinical Characteristics of 17 Hospitalized Patients with 2019 Novel Coronavirus Infections Outside Wuhan, China, medRxiv, (2020).
5.
C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu, X. Gu, Z. Cheng, T. Yu, J. Xia, Y. Wei, W. Wu, X. Xie, W. Yin, H. Li, M. Liu, Y. Xiao, H. Gao, L. Guo, J. Xie, G. Wang, R. Jiang, Z. Gao, Q. Jin, J. Wang, B. Cao, Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, The Lancet, 395 (2020) 497-506.
6.
W.-j. Guan, Z.-y. Ni, Y. Hu, W.-h. Liang, C.-q. Ou, J.-x. He, L. Liu, H. Shan, C.-l. Lei, D.S.C. Hui, B. Du, L.-j. Li, G. Zeng, K.-Y. Yuen, R.-c. Chen, C.-l. Tang, T. Wang, P.-y. Chen, J. Xiang, S.-y. Li, J.-l. Wang, Z.-j. Liang, Y.-x. Peng, L. Wei, Y. Liu, Y.-h. Hu, P. Peng, J.-m. Wang, J.-y. Liu, Z. Chen, G. Li, Z.-j. Zheng, S.-q. Qiu, J. Luo, C.-j. Ye, S.-y. Zhu, N.-s. Zhong, Clinical characteristics of 2019 novel coronavirus infection in China, medRxiv, (2020).
7.
N. Chen, M. Zhou, X. Dong, J. Qu, F. Gong, Y. Han, Y. Qiu, J. Wang, Y. Liu, Y. Wei, J.a. Xia, T. Yu, X. Zhang, L. Zhang, Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study, The Lancet, 395 (2020) 507-513.
8.
A. Wu, Y. Peng, B. Huang, X. Ding, X. Wang, P. Niu, J. Meng, Z. Zhu, Z. Zhang, J. Wang, J. Sheng, L. Quan, Z. Xia, W. Tan, G. Cheng, T. Jiang, Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China, Cell Host Microbe, (2020).
9.
R.C. A. R. Fehr, S. Perlman,, Middle East Respiratory Syndrome:Emergence of a Pathogenic Human Coronavirus, Annu Rev Med 68, 387-399 (2017).
10.
X.Y. Ge, Li, J.L., Yang, X.L., Chmura, A.A., Zhu, G.,Epstein, J.H., Mazet, J.K., Hu, B., Zhang, W., Peng,C., et al. , Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor., Nature 503, 535–538 (2013).
11.
M. Hoffmann, H. Kleine-Weber, N. Krüger, M. Müller, C. Drosten, S. Pöhlmann, The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells, bioRxiv, (2020).
12.
C. Fan, K. Li, Y. Ding, W.L. Lu, J. Wang, ACE2 Expression in Kidney and Testis May Cause Kidney and Testis Damage After 2019-nCoV Infection, medRxiv, (2020).
13.
R. Channappanavar, C. Fett, M. Mack, P.P. Ten Eyck, D.K. Meyerholz, S. Perlman, Sex-Based Differences in Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Infection, The Journal of Immunology, 198 (2017) 4046-4053.
14.
J. Karlberg, D.S. Chong, W.Y. Lai, Do men have a higher case fatality rate of severe acute respiratory syndrome than women do?, Am J Epidemiol, 159 (2004) 229-231.
15.
Z. Li, M. Wu, J. Guo, J. Yao, X. Liao, S. Song, M. Han, J. Li, G. Duan, Y. Zhou, X. Wu, Z. Zhou, T. Wang, M. Hu, X. Chen, Y. Fu, C. Lei, H. Dong, Y. Zhou, H. Jia, X. Chen, J. Yan, Caution on Kidney Dysfunctions of 2019-nCoV Patients, medRxiv, (2020).
16.
W.H. Ding YQ, Shen H, Li ZG, Geng J, Han HX, Cai JJ, Li X, Kang, W.D. W, Lu YD, Wu DH, He L, Yao KT, The clinical pathology of severe acute respiratory syndrome (SARS): a report from China., J Pathol, 2003, 200:282–289 (2003).
17.
Z.L. Lang ZW, Zhang SJ, Meng X, Li JQ, Song CZ, Sun L, Zhou YS, Dwyer DE, A clinicopathological study of three cases of severe acute respiratory syndrome (SARS). , Pathology, 2003, 35:526–531 (2003).
18.
C.P. Chong PY, Ling AE, Franks TJ, Tai DY, Leo YS, Kaw GJ,, C.K. Wansaicheong G, Ean Oon LL, Teo ES, Tan KB, Nakajima, S.T. N, Travis WD, Analysis of deaths during the severe acute respiratory syndrome (SARS) epidemic in Singapore: challenges in determining a SARS diagnosis., Arch Pathol Lab Med, 2004,128:195–204 (2004).
19.
T.W. Chu KH, Tang CS, Lam MF, Lai FM, To KF, Fung KS, Tang HL, Yan WW, Chan HW, Lai TS, Tong KL, Lai KN, Acute renal impairment in coronavirus-associated severe acute respiratory syndrome., Kidney Int, 2005, 67:698–705 (2005).
20.
H.P. Wu VC, Lin WC, Huang JW, Tsai HB, Chen YM, Wu KD, and the SARS Research Group of the National Taiwan, Acute renal failure in SARS patients: more than rhabdomyolysis. , Nephrol Dial Transplant 2004, 19:3180–3182 (2004).
21.
Q.L. Xu J, Chi X, Yang J, Wei X, Gong E, Peh S, Gu J: Orchitis, a complication of severe acute respiratory syndrome (SARS). Biol Reprod, 2006, 74:410–416 (2006).
22.
S. Wan, Q. Yi, S. Fan, J. Lv, X. Zhang, L. Guo, C. Lang, Q. Xiao, K. Xiao, Z. Yi, M. Qiang, J. Xiang, B. Zhang, Y. Chen, Characteristics of lymphocyte subsets and cytokines in peripheral blood of 123 hospitalized patients with 2019-nCoV pneumonia (NCP), medRxiv, (2020).
23.
T. Tanaka, M. Narazaki, T. Kishimoto, IL-6 in inflammation, immunity, and disease, Cold Spring Harb Perspect Biol, 6 (2014) a016295.
24.
A. Shimabukuro-Vornhagen, P. Godel, M. Subklewe, H.J. Stemmler, H.A. Schlosser, M. Schlaak, M. Kochanek, B. Boll, M.S. von Bergwelt-Baildon, Cytokine release syndrome, J Immunother Cancer, 6 (2018) 56.
25.
W.A. Wong RSM, To KF, Lee N, Lam CWK, Wong CK, Chan PKS,, Y.L. Margaret HLN, Hui DS, Tam JS, Cheng G, Sung JJY, Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis., BMJ, 2003, 326:1358–1362 (2003).
26.
D.R. Zhan J, Tang J, Zhang B, Tang Y, Wang JK, Li F, Anderson VM, McNutt MA, Gu J, The spleen as a target in severe acute respiratory syndrome. , FASEB J 2006, 20:2321–2328 (2006).
27.
J. Chen, Y.F. Lau, E.W. Lamirande, C.D. Paddock, J.H. Bartlett, S.R. Zaki, K. Subbarao, Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection, J Virol, 84 (2010) 1289-1301.
28.
D. Wrapp, N. Wang, K.S. Corbett, J.A. Goldsmith, C.-L. Hsieh, O. Abiona, B.S. Graham, J.S. McLellan, Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation, Science, (2020) eabb2507.
29.
H.C. Tong Meng, Hao Zhang, Zijian Kang, Da Xu, Haiyi Gong, Jing Wang, Zifu Li, Xingang Cui, Huji Xu, Haifeng Wei, Xiuwu Pan, Rongrong Zhu, Jianru Xiao, Wang Zhou, Liming Cheng, Jianmin Liu, The insert sequence in SARS-CoV-2 enhances spike protein cleavage by TMPRSS, bioRxiv, (2020).
30.
G.T. Marzi A, Simmons G, Moller P, Rennekamp AJ, Krumbiegel M, Geier M, Eisemann J, Turza N, Saunier B, Steinkasserer A,, B.P. Becker S, Hofmann H, Pohlmann S, DC-SIGN and DCSIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus., J Virol, 2004, 78:12090–12095 (2004).
31.
T.S. Jeffers SA, Gillim-Ross L, Hemmila EM, Achenbach JE,, T.J.W. Babcock GJ, Thackray LB, Young MD, Mason RJ,, W.D. Ambrosino DM, Demartini JC, Holmes KV, CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus, Proc Natl Acad Sci USA 2004, 101:15748–15753 (2004).
Table 1
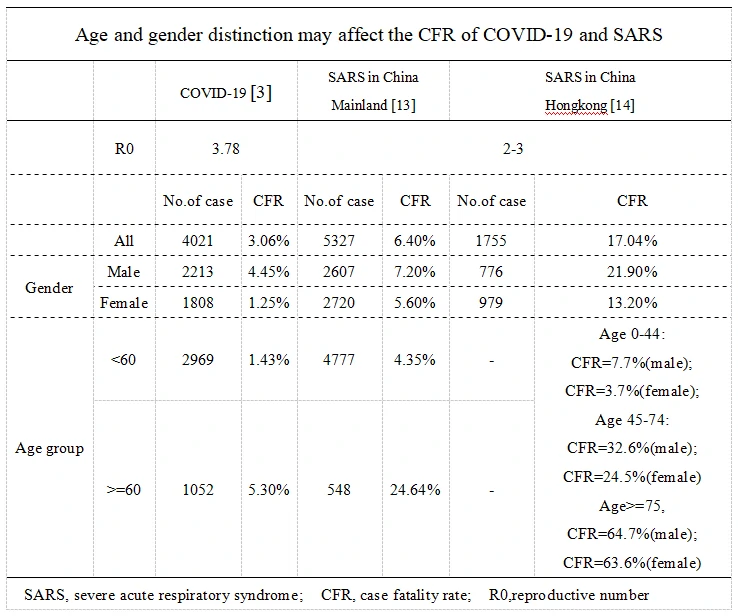 Table 2
Table 2
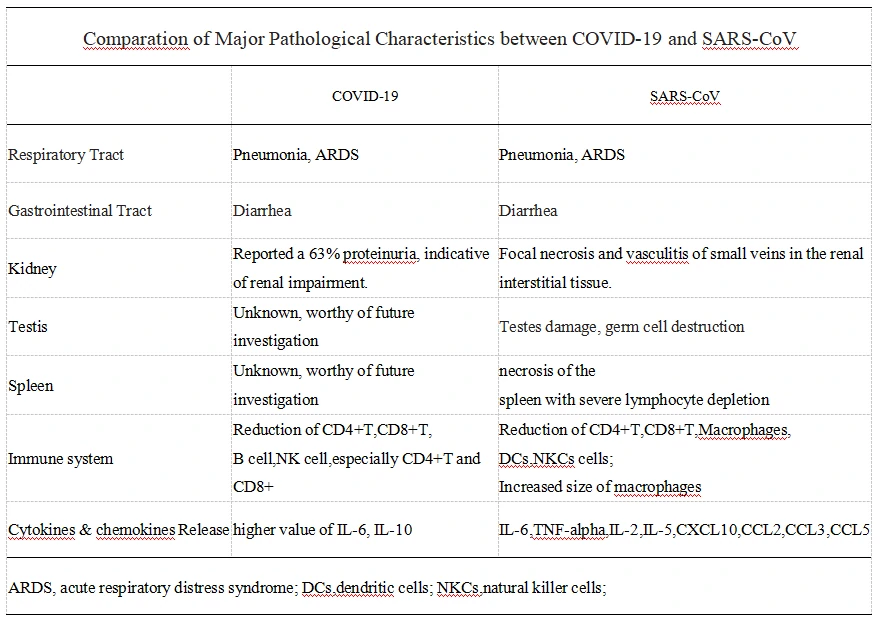
Figure 1. ACE2, L-SIGN(CLEC4M), DC-SIGN(CD209) Protein & RNA expression in organs in Protein Atlas
A. ACE2, L-SIGN(CLEC4M), DC-SIGN(CD209) gene expression in organs classified by gender;
B. ACE2 (the Co-receptor of COVID-19 and SARS-CoV) Protein & RNA expression in organs in Protein Atlas;
C. L-SIGN(CLEC4M, alternative SARS-CoV receptors) Protein & RNA expression in organs in Protein Atlas;
D. DC-SIGN(CD209, alternative SARS-CoV receptors) Protein & RNA expression in organs in Protein Atlas;
Download Picture
Collection of COVID-19 landscape knowledge base
Viral vector-based vaccine; DNA-based vaccine; RNA based vaccine - A landscape for vaccine technology against infectious disease, COVID-19 and tumor.COVID-19 landscape Knowledge Base
Landscape Coronavirus Disease 2019 test (COVID-19 test) in vitro -- A comparison of PCR vs Immunoassay vs Crispr-Based test






