Landscape Coronavirus Disease 2019 test (COVID-19 test) in vitro
-- A comparison of PCR vs Immunoassay vs Crispr-Based test
 Full-text Download
Full-text Download
Author:
Huajun Bai1 Xiaolong Cai1, 2* Xiaoyan Zhang1
1. R&D Center, GeneMedi Co.Ltd., Shanghai, P.R. China (www.genemedi.net)
2. Hanbio Research Center, Hanbio Tech Co. Ltd., Shanghai, P.R. China (www.hanbio.net)
Abstract:
The outbreak of COVID-19, caused by 2019 novel coronavirus (2019-nCoV), has been a global public health threat and caught the worldwide concern. Scientists throughout the world are sparing all efforts to explore strategies for the determination of the 2019-nCoV virus and diagnosis of COVID-19 rapidly. Several assays are developed for COVID-19 test , including RT-PCR, coronavirus antigens-based immunoassays, and CRISPR-based strategies (Cas13a or Cas12a), etc. Different assays have their advantages and drawbacks, and people should choose the most suitable assay according to their demands. Here, we make a brief introduction about these assays and give a simple overview of them, hoping to help doctors and researchers to select the most suitable assay for the Coronavirus Disease 2019 test (COVID-19 test) .
Background
In Dec of 2019, one kind of novel viral pneumonia broke out in Wuhan of China and aroused worldwide concern. This virus was temporarily named as 2019 novel coronavirus (2019-nCoV) by the World Health Organization (WHO) on Jan 7th 2020 [1]. Then, this virus was re-termed as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) based on its sequence similarity to that of 2002-2003 SARS, and the disease it caused was called Coronavirus Disease 2019 (COVID-19) by WHO on Jan 12th, 2020.
SARS-CoV-2 is 60~200nm in diameter and encapsidates a large single-stranded RNA virus (26-32kb) with many spikes on the virus capsid (Fig. 1A). Several characteristic genes typical for coronaviruses (ORF) are observed in the genomes of SARS-CoV (Fig. 1B), such as spike (S), envelope (E), and nucleocapsid (N). Among them, the receptor-binding domain (RBD) of Spike subunit 1 (S1) is indispensable for the viral infection. To date, the mechanisms about how SARS-CoV transduces human cells have not been completely elucidated yet. Whereas a report shows SARS-CoV 2 infects human with similar processes to SARS by binding to the angiotensin-converting enzyme 2 (ACE2) receptor of target cells [2-4], such as respiratory epithelial cells. Once humans are infected with SARS-CoV-2, they will have the following symptoms: ① having fever and feeling fatigue systematically; ② sneezing, runny nose, sore throat, dry cough, and shortness of breath in the respiratory system; ③ diseased function in kidney; ④ diarrhea in intestines; ⑤ deceased white blood cells.
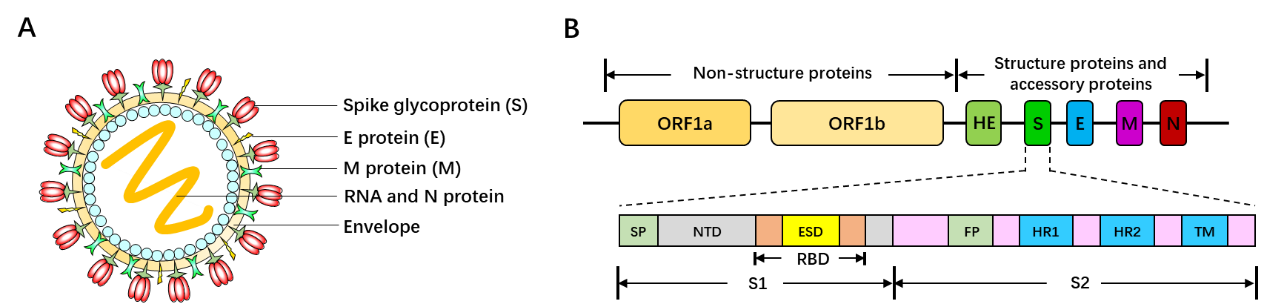
Figure 1. SARS-CoV-2 capsid structure and genome map. (A) Three-dimensional structure diagram of SARS-CoV-2. (B) Genome organization of SARS-CoV-2 [5]. ORF: open reading frame. E: envelope. M: membrane. N: nucleocapsid. HR1: heptad repeat 1. HR2: heptad repeat 2. SP: signal peptide. NTD: N-terminal domain. RBD: receptor binding domain. S: spike. S1: subunit 1. S2: subunit 2. TM: transmembrane domain.
It has been reported that person-to-person transmission from infected COVID-19 patients is really rapidly [5-7]. Although China government is basically in control of COVID-19 pathophoresis by separating patients from normal, forcing citizens to wear masks, and controlling traffic, now the viral pneumonia is globally threatening the health of people all over the world, especially in Europe and North America. Since no specific therapeutic drugs or vaccines are available for patients with COVID-19, it is really necessary and noteworthy to determine whether the patient is infected early and separate the infected patients from the healthy population immediately to avoid the widespread of SARS-CoV-2. To date, there are several strategies for COVID-19 test: ① real-time PCR (RT-PCR) method; ② immunoassay; ③ Crispr-Cas13a (SHERLOCK)-based test.
RT-PCR (Real time -PCR) in Coronavirus Disease 2019 (COVID-19) test
1) Principles for diagnostics

Figure 2. Principle of qRT-PCR.
2) Advantages and disadvantages
Advantages:
① High sensitivity.
② Easy to be operated on a large scale.
Disadvantages:
① Professional technician and special apparatus are required to perform RT-PCR experiments and analyze the data, which costs so much.
② The standard positive control affects the experimental accuracy, and a false negative result may occur due to improper handling.
3) Optional Targets, primers, and probes from different departments
Different departments have their own systems for the determination of COVID-19 by targeting various open reading frames of SARS-CoV-2, which are shown as follows. Primers and probes of Orf1ab and N (nucleocapsid gene) are commonly used for COVID-19 test in China. Three pairs of primers and probes of N (nucleocapsid gene, N1, N2, N3) are applied in the US CDC for COVID-19. And primers and probes of E (envelope gene) are utilized for COVID-19 test in Europe.
| Organisation | Target | Forward primer (5’-3’) | Reverse primer (5’-3’) | Probe (5’-3’) |
University of Hong Kong, Beijing Center for Disease Prevention and Control, Capital Medical University Beijing Research Center for Preventive Medicine | orf1b | TGGGGYTTTACRGGTAACCT (Forward; Y=C/T, R=A/G) | AACRCGCTTAACAAAGCACTC (Reverse; R=A/G) | TAGTTGTGATGCWATCATGACTAG (Probe in 5’-FAM/ZEN/3’-IBFQ format; W=A/T) |
| N | TAATCAGACAAGGAACTGATTA | CGAAGGTGTGACTTCCATG | FAM-GCAAATTGTGCAATTTGCGG-IBFQ | |
Charité Virology, Berlin, Germany; Olfert Landt, Tib-Molbiol, Berlin, Germany; Erasmus MC, Rotterdam, The Netherlands; Public Health England, London | RdRP | GTGARATGGTCATGT GTGGCGG | CARATGTTAAASACA CTATTAGCATA | FAM-CAGGTGGA ACCTCATCAGGA GATGCBBQ |
| E gene | ACAGGTACGTTAATA GTTAATAGCGT | ATATTGCAGCAGTAC GCACACA | FAM-ACACTAGC CATCCTTACTGC GCTTCGBBQ | |
| N | CACATTGGCACCCGC AATC | GAGGAACGAGAAGA GGCTTG | FAM-ACTTCCTCA AGGAACAACATT GCCABBQ | |
The US Centers for Disease Control and Prevention (CDC), Integrated DNA Technologies | N1 | GACCCCAAAATCAGC GAAAT | TCTGGTTACTGCCAG TTG AATCTG | FAM-ACCCCGCA T TACGTTTGGTGG ACC-BHQ1 |
| N2 | TTACAAACATTGGCCGCAAA | GCGCGACATTCCGAAGAA | FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ1 | |
| N3 | GGGAGCCTTGAATACACCAAAA | TGTAGCACGATTGCAGCATTG | FAM-AYCACATTGGCACCCGCAATCCTG-BHQ1 | |
| RNAse P | AGATTTGGACCTGCGAGCG | GAGCGGCTGTCTCCACAAGT | FAM–TTCTGACCTGAAGGCTCTGCGCG–BHQ-1 |
Table 1. Optional Targets, primers, and probes from different departments
Immunoassay in Coronavirus Disease 2019 (COVID-19) test
1) Point-of-care lateral flow test
A. Principles for Diagnostics
Once entry into host cells, SARS-CoV-2 will provoke the adaptive immune system in the body of the host. 3-6 days post-infection, IgM antibody will be produced, while IgG antibody will be generated 8 days post-infection. Thus, these antibodies against SARS-CoV-2 can be detected in patient’s blood. Moreover, IgM tends to be an indicator of the recent exposure of virus, while IgG indicates earlier virus infection. The IgM and IgG can be determined based on highly specific antibody-antigen interactions. In brief, add 10-15μl blood specimens and 70μl sample dilution buffer to the sample port. When the blood specimen flows through the SARS-CoV-2 recombinant antigen (receptor binding domain of SARS-CoV-2 spike protein) labeled by 40nm gold nanoparticle (AuNP) colloids via chromatographic lateral flow, a complex of IgM-antigen-AuNP/IgG-antigen-AuNP will form. Then this complex continues to move on and passes through the coated mouse anti-human IgM/G antibody at M/G coating line, they will form a double antibody sandwich colloidal gold complex, showing a color band at the coating line (red/purple). The excessive fluid continues to flow through the Control line, Rabbit IgG conjugated AuNP will interact with Goat anti-Rabbit IgG antibody, exhibiting a color band at the control line [8]. This point-of-care lateral flow test utilizes a colloidal gold as an indicator to determine the IgM/G against SARS-CoV-2, which is a reliable and visual index for the rapid screening of SARS-CoV-2 carriers (Fig. 3).
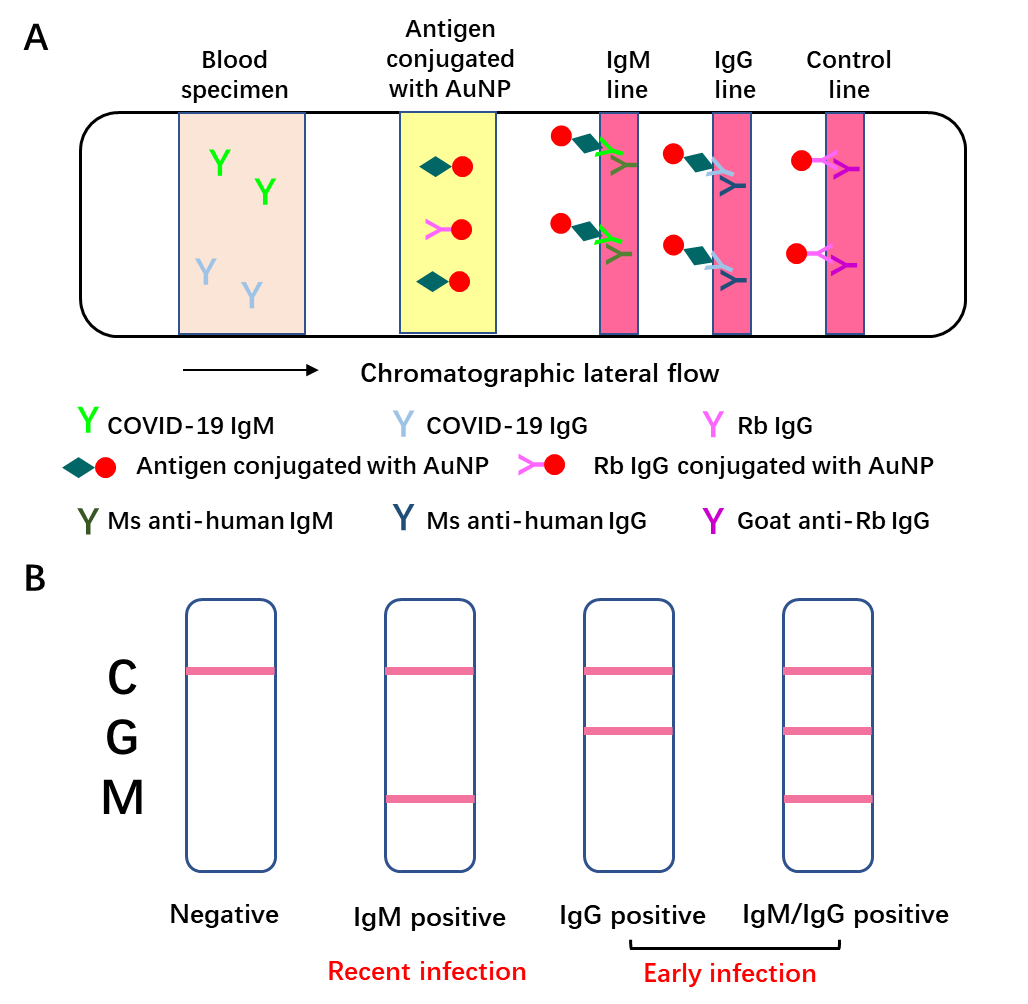
Figure 3. Schematic illustration of Point-of-care lateral flow immunoassay [8].
A: Schematic diagram of rapid SARS-CoV-2 IgM-IgG combined antibody detection device;
B: An illustration of different testing results. C: control line, G: IgG line, M: IgM line.
B. Advantages and disadvantages
Advantages:
① Easy to use and operate on a large scale.
② No requirement for additional equipment.
③ High sensitivity (88.66%) and specificity (90.63%).
④ Cost little time, less than 15 minutes.
Disadvantages:
Long window period. It’s difficult to detect the early infections, for IgM antibody could only be detected in patient’s blood 3-6 days post-infection, while IgG can be detected 8 days post-infection.
C. Optional Targets
The targets of point-of-care lateral flow tests are human antibodies against SARS-CoV-2 antigens. SARS-CoV-2 mainly has four structural proteins with great genetic similarity to those of SARS-CoV, including S (76.0%), E (94.7%), M (90.1%), and N (90.6%). Based on the antigen epitope analysis of SARS-CoV and SARS-CoV-2 [9], N protein and S protein are expected to be potential antigen targets with a large number of epitopes for the induction of both T cell response and B cell response, which might show promising prospects for vaccine development and induction of long-term immune responses. Additionally, several structural shreds of evidence show S protein is crucial for the entry of SARS-CoV-2 into host cells [2-4, 10] and administration of antibody against SARS-CoV S protein blocks SARS-CoV-2 S mediated entry into cells, which might give a blueprint for COVID-19 vaccine development targeting S protein epitopes [3].
2) ELISA and the other immunoassay in Coronavirus Disease 2019 (COVID-19) test
Besides the above Point-of-care lateral flow test, ELlSA also can be utilized to quantify SARS-CoV-2 antigens via the double antibody sandwich method. Briefly, coat the plate with capture antibody, which can interact with antigens and pull the antigens down once sample is added. Then detection antibody, conjugated with biotin or fluorophores, is added to bind to the other epitope of antigens. Thus, antigen can be detected with capture antibody and detection antibody and labeled by biotin or fluorophores. Additionally, when SARS-CoV-2 infects humans, the immune system will generate a pile of cytokines, including IL2, IL7, IL10, GSCF, IP10, MCP1, MIP1A, and TNFα [11]. These cytokines can also be detected by ELISA or other immunoassays, such as Western blot or immunostaining.
3) Core for COVID-19 immunoassay: recombinant 2019nCoV antigens
All of these immunoassays for COVID-19 test are based on the antigens of 2019nCoV. To date, GeneMedi has produced many recombinant antigens, including Nucleocapsid (N protein), Spike protein (S protein, S1+S2 ECD), Spike protein (S1 protein), spike RBD protein, Envelope (E protein), 3C-like Proteinase. These recombinant antigens are available and ready-to-use on GeneMedi: https://www.genemedi.net/i/recombinant-2019-ncov-antigens-reagents
CRISPR-based tools for Coronavirus Disease 2019 (COVID-19) test
1) Principles for Diagnostics
As RT-PCR assay for COVID-19 requires several reagents, specific instruments and complicated processes, CRISPR-based tools have been developed to test COVID-19 within 30 minutes, which is really easy and simple to operated and saves a lot of time.
A. CRISPR-Cas13a (SHERLOCK)
Based on CRISPR (clustered regularly interspaced short palindromic repeats) technology, single-effector RNA-guided ribonucleases (RNases), such as Cas13a (previously known as C2c2) [12, 13], can be activated to engage in “collateral” cleavage of nearby nontargeted RNAs once recognizing its RNA targets. Recombinase polymerase amplification (RPA) can efficiently amplify DNA from single-molecule to 1012 level at a low isothermal state with no need to break the double-stranded structure of DNA [14]. Combined with RPA and T7 RNA polymerase, the CRISPR-Cas13a system can detect RNA rapidly with attomolar (10-18) sensitivity. This technology can be applied for real-time detection of the presence of target RNAs in vitro with signal amplification by non-specific collateral cleavage of nearby nontargeted reporter RNA (Fig. 4) [15]. Therefore, this system is called Specific High-Sensitivity Enzymatic Reporter UnLOCKing (SHERLOCK) based on the amplification of nucleic acid and Cas13a-mediated collateral cleavage of a reporter RNA.

Figure 4. General principle of SHERLOCK tool. dsDNA, double-stranded DNA; RPA, recombinase polymerase amplification, RT-RPA, reverse transcriptase–RPA [15].
For COVID-19 determination, two specific gRNAs targeting Orf1ab and S gene are used and the sequences are as follows. The SHERLOCK COVID-19 detection protocol only requires 3 steps: ① 25min incubation for isothermal amplification of the extracted nucleic acid sample with RPA kit; ② 30 min incubation for the determination of pre-amplified viral RNA sequence using Cas13 protein; ③ 2 min incubation for visual read out of the detection result with a commercially-available paper dipstick.
| S gene crRNA | 5’-GAUUUAGACUACCCCAAAAACGAAGGGGACUAAAACGCAGCACCAGCUGUCCAACCUGAAGAAG-3’ |
| Orf1ab-crRNA | 5’-GAUUUAGACUACCCCAAAAACGAAGGGGACUAAAACCCAACCUCUUCUGUAAUUUUUAAACUAU-3’ |
| Reporter RNA | 5’-/56-FAM/mArArUrGrGrCmAmArArUrGrGrCmA/3Bio/-3’ |
Table 2. crRNA for SHERLOCK COVID-19 detection
B. CRISPR-based DETECT Lateral Flow Assay
Similarly, another diagnostic tool based on CRISPR is DETECTR (DNA endonuclease-targeted CRISPR trans reporter) system [16, 17]. DETECTR system carry out reverse transcription as well as DNA amplification simultaneously with loop-mediated amplification (RT-LAMP) [18]. The gRNAs are designed to target E gene and N gene of SARS-CoV-2 with the protospacer adjacent motif (PAM) sequence for Cas12 (also known as Cpf1) enzyme [19]. When Cas12 specifically recognizes and binds to the target single-stranded DNA (ssDNA), it will be activated to completely degrade ssDNA molecules non-specifically. DETECTR system is based on the isothermal amplification of target DNA and Cas12-mediated to collateral cleavage the ssDNA probe. The detailed workflow of DETECTR system-based COVID-19 test is shown in Fig. 6 [20], similarly to SHERLOCK system.
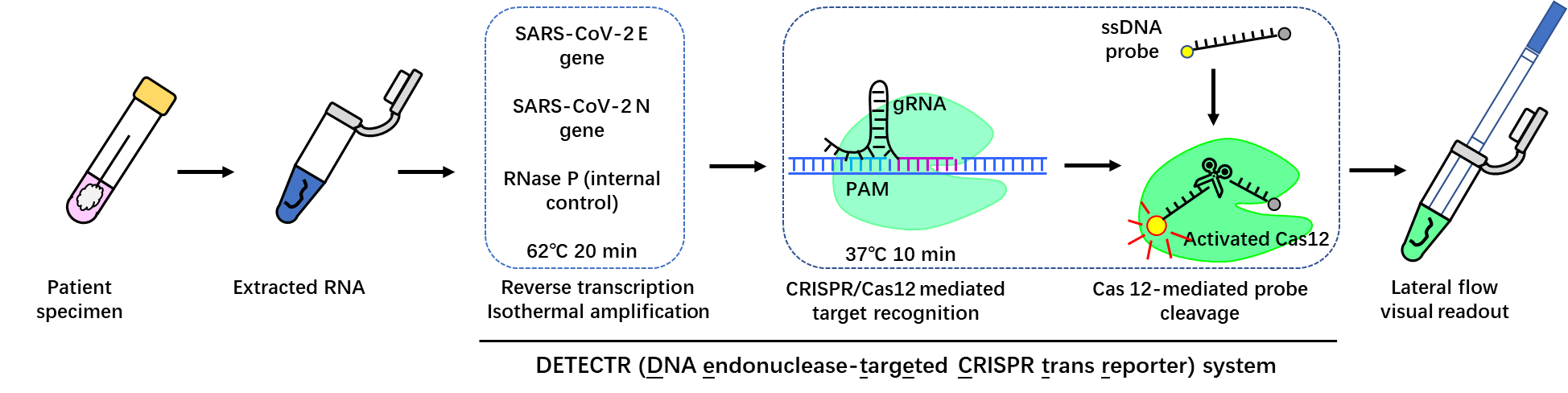
Figure 5. Flow diagram of COVID-19 test based on DETECTR system [20].
| N gene crRNA | 5'-CACAATTTGCCCCCAGCGCTTCAGCGTTCTTCGG-3' |
| E gene crRNA | 5'-CTTGCTTTCGTGGTATTCTTGCTAGTTACACTAG-3' |
| Reporter | 5’-/56-FAM/TTATTATT/3Bio/-3’, IDT |
Table 3. crRNA for DETECTR system COVID-19 detection
2) Advantages and disadvantages
Advantages:
① Easy to use and operate in a large scale.
② No requirement of additional equipment.
③ High specificity and sensitivity. Both of the two molecular diagnostic technologies, SHERLOCK and DETECTR, can be used to detect specific RNA/DNA at the attomolar level [16, 17].
④ Cost little time, within 1h.
⑤ Can detect early infection of SARS-CoV-2.
Disadvantages:
There may exist some off-target effects.
Summary
Among the three methods for COVID-19 test, IgM/IgG point-of-care lateral flow test is the simplest and most rapid regardless of professional skill, and cost the least time with no requirement of additional instruments. Based on the detection of viral RNA, CRISPR-based tests are simpler and easier than RT-PCR test with no need for bulky instruments and complicated operations.
| Methods | RT-PCR | Immunoassay based point-of-care lateral flow test | CRISPR-based test |
| Specificity | High | High | High | Consumption time | 4-6 hours | Within 15 min | Within 1 hour |
| Bulky instrumentation | Required | No requirement | No requirement |
| Disadvantage | False negative result may occur due to improper handling | Long term needed for antibody generation after infection | Off-targets may exist |
| Cost | High | Low | Low |
| Target | Different departments have various targets, such as Orf1ab, N gene, E gene | IgM/IgG against SARS-CoV-2 | SHERLOCK system: Orf1ab, S gene; DETECTR system: E gene and N gene. |
| Sample | Urine, blood, saliva, specimen from respiratory, etc. | Blood | Urine, blood, saliva, etc. |
Table 4 A summary and comparison of different strategies for COVID-19 test.
Acknowledgement
Tribute to the medical staff fighting against the COVID-19 all over the world. Thanks for the helps from our colleagues from Hanbio Tech Co. Ltd., Shanghai, P.R. China (www.hanbio.net) and GeneMedi Co.Ltd., Shanghai, P.R. China (www.genemedi.net) respectively.
Reference
1.
L.E.a.V.D.M. Gralinski, Return of the Coronavirus: 2019-nCoV. , Viruses, 2020. 12(2). (2020).
2.
J.L. V. M. Corman, M. Witzenrath, Coronaviruses as the cause of respiratory infections, Internist (Berl) 60, 1136-1145 (2019).
3.
Y. Yang, Q. Lu, M. Liu, Y. Wang, A. Zhang, N. Jalali, N. Dean, I. Longini, M.E. Halloran, B. Xu, X. Zhang, L. Wang, W. Liu, L. Fang, Epidemiological and clinical features of the 2019 novel coronavirus outbreak in China, medRxiv, (2020).
4.
J. Li, S. Li, Y. Cai, Q. Liu, X. Li, Z. Zeng, Y. Chu, F. Zhu, F. Zeng, Epidemiological and Clinical Characteristics of 17 Hospitalized Patients with 2019 Novel Coronavirus Infections Outside Wuhan, China, medRxiv, (2020).
5.
C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu, X. Gu, Z. Cheng, T. Yu, J. Xia, Y. Wei, W. Wu, X. Xie, W. Yin, H. Li, M. Liu, Y. Xiao, H. Gao, L. Guo, J. Xie, G. Wang, R. Jiang, Z. Gao, Q. Jin, J. Wang, B. Cao, Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, The Lancet, 395 (2020) 497-506.
6.
W.-j. Guan, Z.-y. Ni, Y. Hu, W.-h. Liang, C.-q. Ou, J.-x. He, L. Liu, H. Shan, C.-l. Lei, D.S.C. Hui, B. Du, L.-j. Li, G. Zeng, K.-Y. Yuen, R.-c. Chen, C.-l. Tang, T. Wang, P.-y. Chen, J. Xiang, S.-y. Li, J.-l. Wang, Z.-j. Liang, Y.-x. Peng, L. Wei, Y. Liu, Y.-h. Hu, P. Peng, J.-m. Wang, J.-y. Liu, Z. Chen, G. Li, Z.-j. Zheng, S.-q. Qiu, J. Luo, C.-j. Ye, S.-y. Zhu, N.-s. Zhong, Clinical characteristics of 2019 novel coronavirus infection in China, medRxiv, (2020).
7.
N. Chen, M. Zhou, X. Dong, J. Qu, F. Gong, Y. Han, Y. Qiu, J. Wang, Y. Liu, Y. Wei, J.a. Xia, T. Yu, X. Zhang, L. Zhang, Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study, The Lancet, 395 (2020) 507-513.
8.
A. Wu, Y. Peng, B. Huang, X. Ding, X. Wang, P. Niu, J. Meng, Z. Zhu, Z. Zhang, J. Wang, J. Sheng, L. Quan, Z. Xia, W. Tan, G. Cheng, T. Jiang, Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China, Cell Host Microbe, (2020).
9.
R.C. A. R. Fehr, S. Perlman,, Middle East Respiratory Syndrome:Emergence of a Pathogenic Human Coronavirus, Annu Rev Med 68, 387-399 (2017).
10.
X.Y. Ge, Li, J.L., Yang, X.L., Chmura, A.A., Zhu, G.,Epstein, J.H., Mazet, J.K., Hu, B., Zhang, W., Peng,C., et al. , Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor., Nature 503, 535–538 (2013).
11.
M. Hoffmann, H. Kleine-Weber, N. Krüger, M. Müller, C. Drosten, S. Pöhlmann, The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells, bioRxiv, (2020).
12.
C. Fan, K. Li, Y. Ding, W.L. Lu, J. Wang, ACE2 Expression in Kidney and Testis May Cause Kidney and Testis Damage After 2019-nCoV Infection, medRxiv, (2020).
13.
R. Channappanavar, C. Fett, M. Mack, P.P. Ten Eyck, D.K. Meyerholz, S. Perlman, Sex-Based Differences in Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Infection, The Journal of Immunology, 198 (2017) 4046-4053.
14.
J. Karlberg, D.S. Chong, W.Y. Lai, Do men have a higher case fatality rate of severe acute respiratory syndrome than women do?, Am J Epidemiol, 159 (2004) 229-231.
15.
Z. Li, M. Wu, J. Guo, J. Yao, X. Liao, S. Song, M. Han, J. Li, G. Duan, Y. Zhou, X. Wu, Z. Zhou, T. Wang, M. Hu, X. Chen, Y. Fu, C. Lei, H. Dong, Y. Zhou, H. Jia, X. Chen, J. Yan, Caution on Kidney Dysfunctions of 2019-nCoV Patients, medRxiv, (2020).
16.
W.H. Ding YQ, Shen H, Li ZG, Geng J, Han HX, Cai JJ, Li X, Kang, W.D. W, Lu YD, Wu DH, He L, Yao KT, The clinical pathology of severe acute respiratory syndrome (SARS): a report from China., J Pathol, 2003, 200:282–289 (2003).
17.
Z.L. Lang ZW, Zhang SJ, Meng X, Li JQ, Song CZ, Sun L, Zhou YS, Dwyer DE, A clinicopathological study of three cases of severe acute respiratory syndrome (SARS). , Pathology, 2003, 35:526–531 (2003).
18.
C.P. Chong PY, Ling AE, Franks TJ, Tai DY, Leo YS, Kaw GJ,, C.K. Wansaicheong G, Ean Oon LL, Teo ES, Tan KB, Nakajima, S.T. N, Travis WD, Analysis of deaths during the severe acute respiratory syndrome (SARS) epidemic in Singapore: challenges in determining a SARS diagnosis., Arch Pathol Lab Med, 2004,128:195–204 (2004).
19.
T.W. Chu KH, Tang CS, Lam MF, Lai FM, To KF, Fung KS, Tang HL, Yan WW, Chan HW, Lai TS, Tong KL, Lai KN, Acute renal impairment in coronavirus-associated severe acute respiratory syndrome., Kidney Int, 2005, 67:698–705 (2005).
20.
H.P. Wu VC, Lin WC, Huang JW, Tsai HB, Chen YM, Wu KD, and the SARS Research Group of the National Taiwan, Acute renal failure in SARS patients: more than rhabdomyolysis. , Nephrol Dial Transplant 2004, 19:3180–3182 (2004).
Collection of COVID-19 landscape knowledge base
Viral vector-based vaccine; DNA-based vaccine; RNA based vaccine
- A landscape for vaccine technology against infectious disease, COVID-19 and tumor.
An Insight of comparison between COVID-19 (2019-nCoV disease) and SARS in pathology and pathogenesis
COVID-19 landscape Knowledge Base






