CRISPR-based tools for Coronavirus Disease 2019 (COVID-19) test
 Full-text Download
Full-text Download
Author:
Huajun Bai1 Xiaolong Cai1, 2* Xiaoyan Zhang1
1. R&D Center, GeneMedi Co.Ltd., Shanghai, P.R. China (www.genemedi.net)
2. Hanbio Research Center, Hanbio Tech Co. Ltd., Shanghai, P.R. China (www.hanbio.net)
Abstract:
The outbreak of COVID-19, caused by 2019 novel coronavirus (2019-nCoV), has been a global public health threat and caught the worldwide concern. Scientists throughout the world are sparing all efforts to explore strategies for the determination of the 2019-nCoV virus and diagnosis of COVID-19 rapidly. Several assays are developed for COVID-19 test , including RT-PCR, coronavirus antigens-based immunoassays, and CRISPR-based strategies (Cas13a or Cas12a), etc. Different assays have their advantages and drawbacks, and people should choose the most suitable assay according to their demands. Here, we make a brief introduction about these assays and give a simple overview of them, hoping to help doctors and researchers to select the most suitable assay for the Coronavirus Disease 2019 test (COVID-19 test) .
CRISPR-based tools for Coronavirus Disease 2019 (COVID-19) test
1) Principles for Diagnostics
As RT-PCR assay for COVID-19 requires several reagents, specific instruments and complicated processes, CRISPR-based tools have been developed to test COVID-19 within 30 minutes, which is really easy and simple to operated and saves a lot of time.
A. CRISPR-Cas13a (SHERLOCK)
Based on CRISPR (clustered regularly interspaced short palindromic repeats) technology, single-effector RNA-guided ribonucleases (RNases), such as Cas13a (previously known as C2c2) [12, 13], can be activated to engage in “collateral” cleavage of nearby nontargeted RNAs once recognizing its RNA targets. Recombinase polymerase amplification (RPA) can efficiently amplify DNA from single-molecule to 1012 level at a low isothermal state with no need to break the double-stranded structure of DNA [14]. Combined with RPA and T7 RNA polymerase, the CRISPR-Cas13a system can detect RNA rapidly with attomolar (10-18) sensitivity. This technology can be applied for real-time detection of the presence of target RNAs in vitro with signal amplification by non-specific collateral cleavage of nearby nontargeted reporter RNA (Fig. 4) [15]. Therefore, this system is called Specific High-Sensitivity Enzymatic Reporter UnLOCKing (SHERLOCK) based on the amplification of nucleic acid and Cas13a-mediated collateral cleavage of a reporter RNA.

Figure 4. General principle of SHERLOCK tool. dsDNA, double-stranded DNA; RPA, recombinase polymerase amplification, RT-RPA, reverse transcriptase–RPA [15].
For COVID-19 determination, two specific gRNAs targeting Orf1ab and S gene are used and the sequences are as follows. The SHERLOCK COVID-19 detection protocol only requires 3 steps: ① 25min incubation for isothermal amplification of the extracted nucleic acid sample with RPA kit; ② 30 min incubation for the determination of pre-amplified viral RNA sequence using Cas13 protein; ③ 2 min incubation for visual read out of the detection result with a commercially-available paper dipstick.
| S gene crRNA | 5’-GAUUUAGACUACCCCAAAAACGAAGGGGACUAAAACGCAGCACCAGCUGUCCAACCUGAAGAAG-3’ |
| Orf1ab-crRNA | 5’-GAUUUAGACUACCCCAAAAACGAAGGGGACUAAAACCCAACCUCUUCUGUAAUUUUUAAACUAU-3’ |
| Reporter RNA | 5’-/56-FAM/mArArUrGrGrCmAmArArUrGrGrCmA/3Bio/-3’ |
Table 2. crRNA for SHERLOCK COVID-19 detection
B. CRISPR-based DETECT Lateral Flow Assay
Similarly, another diagnostic tool based on CRISPR is DETECTR (DNA endonuclease-targeted CRISPR trans reporter) system [16, 17]. DETECTR system carry out reverse transcription as well as DNA amplification simultaneously with loop-mediated amplification (RT-LAMP) [18]. The gRNAs are designed to target E gene and N gene of SARS-CoV-2 with the protospacer adjacent motif (PAM) sequence for Cas12 (also known as Cpf1) enzyme [19]. When Cas12 specifically recognizes and binds to the target single-stranded DNA (ssDNA), it will be activated to completely degrade ssDNA molecules non-specifically. DETECTR system is based on the isothermal amplification of target DNA and Cas12-mediated to collateral cleavage the ssDNA probe. The detailed workflow of DETECTR system-based COVID-19 test is shown in Fig. 6 [20], similarly to SHERLOCK system.
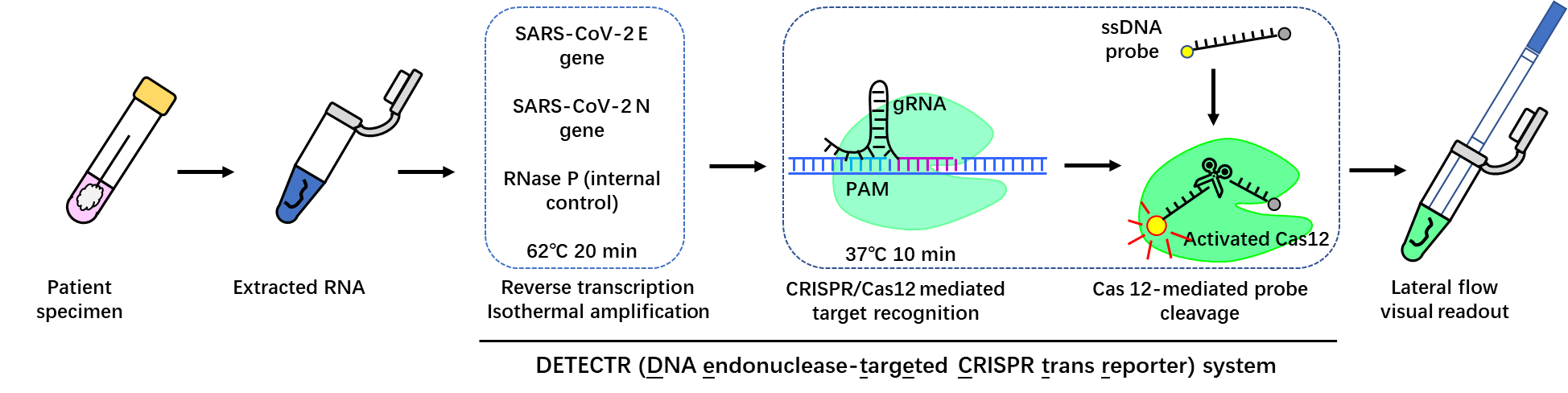
Figure 5. Flow diagram of COVID-19 test based on DETECTR system [20].
| N gene crRNA | 5'-CACAATTTGCCCCCAGCGCTTCAGCGTTCTTCGG-3' |
| E gene crRNA | 5'-CTTGCTTTCGTGGTATTCTTGCTAGTTACACTAG-3' |
| Reporter | 5’-/56-FAM/TTATTATT/3Bio/-3’, IDT |
Table 3. crRNA for DETECTR system COVID-19 detection
2) Advantages and disadvantages
Advantages:
① Easy to use and operate in a large scale.
② No requirement of additional equipment.
③ High specificity and sensitivity. Both of the two molecular diagnostic technologies, SHERLOCK and DETECTR, can be used to detect specific RNA/DNA at the attomolar level [16, 17].
④ Cost little time, within 1h.
⑤ Can detect early infection of SARS-CoV-2.
Disadvantages:
There may exist some off-target effects.
Reference
1.
L.E.a.V.D.M. Gralinski, Return of the Coronavirus: 2019-nCoV. , Viruses, 2020. 12(2). (2020).
2.
J.L. V. M. Corman, M. Witzenrath, Coronaviruses as the cause of respiratory infections, Internist (Berl) 60, 1136-1145 (2019).
3.
Y. Yang, Q. Lu, M. Liu, Y. Wang, A. Zhang, N. Jalali, N. Dean, I. Longini, M.E. Halloran, B. Xu, X. Zhang, L. Wang, W. Liu, L. Fang, Epidemiological and clinical features of the 2019 novel coronavirus outbreak in China, medRxiv, (2020).
4.
J. Li, S. Li, Y. Cai, Q. Liu, X. Li, Z. Zeng, Y. Chu, F. Zhu, F. Zeng, Epidemiological and Clinical Characteristics of 17 Hospitalized Patients with 2019 Novel Coronavirus Infections Outside Wuhan, China, medRxiv, (2020).
5.
C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu, X. Gu, Z. Cheng, T. Yu, J. Xia, Y. Wei, W. Wu, X. Xie, W. Yin, H. Li, M. Liu, Y. Xiao, H. Gao, L. Guo, J. Xie, G. Wang, R. Jiang, Z. Gao, Q. Jin, J. Wang, B. Cao, Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, The Lancet, 395 (2020) 497-506.
6.
W.-j. Guan, Z.-y. Ni, Y. Hu, W.-h. Liang, C.-q. Ou, J.-x. He, L. Liu, H. Shan, C.-l. Lei, D.S.C. Hui, B. Du, L.-j. Li, G. Zeng, K.-Y. Yuen, R.-c. Chen, C.-l. Tang, T. Wang, P.-y. Chen, J. Xiang, S.-y. Li, J.-l. Wang, Z.-j. Liang, Y.-x. Peng, L. Wei, Y. Liu, Y.-h. Hu, P. Peng, J.-m. Wang, J.-y. Liu, Z. Chen, G. Li, Z.-j. Zheng, S.-q. Qiu, J. Luo, C.-j. Ye, S.-y. Zhu, N.-s. Zhong, Clinical characteristics of 2019 novel coronavirus infection in China, medRxiv, (2020).
7.
N. Chen, M. Zhou, X. Dong, J. Qu, F. Gong, Y. Han, Y. Qiu, J. Wang, Y. Liu, Y. Wei, J.a. Xia, T. Yu, X. Zhang, L. Zhang, Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study, The Lancet, 395 (2020) 507-513.
8.
A. Wu, Y. Peng, B. Huang, X. Ding, X. Wang, P. Niu, J. Meng, Z. Zhu, Z. Zhang, J. Wang, J. Sheng, L. Quan, Z. Xia, W. Tan, G. Cheng, T. Jiang, Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China, Cell Host Microbe, (2020).
9.
R.C. A. R. Fehr, S. Perlman,, Middle East Respiratory Syndrome:Emergence of a Pathogenic Human Coronavirus, Annu Rev Med 68, 387-399 (2017).
10.
X.Y. Ge, Li, J.L., Yang, X.L., Chmura, A.A., Zhu, G.,Epstein, J.H., Mazet, J.K., Hu, B., Zhang, W., Peng,C., et al. , Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor., Nature 503, 535–538 (2013).
11.
M. Hoffmann, H. Kleine-Weber, N. Krüger, M. Müller, C. Drosten, S. Pöhlmann, The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells, bioRxiv, (2020).
12.
C. Fan, K. Li, Y. Ding, W.L. Lu, J. Wang, ACE2 Expression in Kidney and Testis May Cause Kidney and Testis Damage After 2019-nCoV Infection, medRxiv, (2020).
13.
R. Channappanavar, C. Fett, M. Mack, P.P. Ten Eyck, D.K. Meyerholz, S. Perlman, Sex-Based Differences in Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Infection, The Journal of Immunology, 198 (2017) 4046-4053.
14.
J. Karlberg, D.S. Chong, W.Y. Lai, Do men have a higher case fatality rate of severe acute respiratory syndrome than women do?, Am J Epidemiol, 159 (2004) 229-231.
15.
Z. Li, M. Wu, J. Guo, J. Yao, X. Liao, S. Song, M. Han, J. Li, G. Duan, Y. Zhou, X. Wu, Z. Zhou, T. Wang, M. Hu, X. Chen, Y. Fu, C. Lei, H. Dong, Y. Zhou, H. Jia, X. Chen, J. Yan, Caution on Kidney Dysfunctions of 2019-nCoV Patients, medRxiv, (2020).
16.
W.H. Ding YQ, Shen H, Li ZG, Geng J, Han HX, Cai JJ, Li X, Kang, W.D. W, Lu YD, Wu DH, He L, Yao KT, The clinical pathology of severe acute respiratory syndrome (SARS): a report from China., J Pathol, 2003, 200:282–289 (2003).
17.
Z.L. Lang ZW, Zhang SJ, Meng X, Li JQ, Song CZ, Sun L, Zhou YS, Dwyer DE, A clinicopathological study of three cases of severe acute respiratory syndrome (SARS). , Pathology, 2003, 35:526–531 (2003).
18.
C.P. Chong PY, Ling AE, Franks TJ, Tai DY, Leo YS, Kaw GJ,, C.K. Wansaicheong G, Ean Oon LL, Teo ES, Tan KB, Nakajima, S.T. N, Travis WD, Analysis of deaths during the severe acute respiratory syndrome (SARS) epidemic in Singapore: challenges in determining a SARS diagnosis., Arch Pathol Lab Med, 2004,128:195–204 (2004).
19.
T.W. Chu KH, Tang CS, Lam MF, Lai FM, To KF, Fung KS, Tang HL, Yan WW, Chan HW, Lai TS, Tong KL, Lai KN, Acute renal impairment in coronavirus-associated severe acute respiratory syndrome., Kidney Int, 2005, 67:698–705 (2005).
20.
H.P. Wu VC, Lin WC, Huang JW, Tsai HB, Chen YM, Wu KD, and the SARS Research Group of the National Taiwan, Acute renal failure in SARS patients: more than rhabdomyolysis. , Nephrol Dial Transplant 2004, 19:3180–3182 (2004).
Collection of COVID-19 landscape knowledge base
Viral vector-based vaccine; DNA-based vaccine; RNA based vaccine
- A landscape for vaccine technology against infectious disease, COVID-19 and tumor.
An Insight of comparison between COVID-19 (2019-nCoV disease) and SARS in pathology and pathogenesis
COVID-19 landscape Knowledge Base






