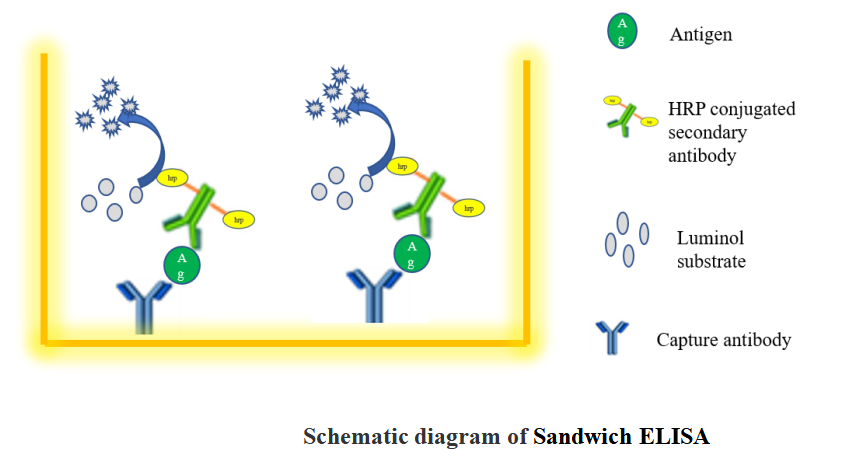
What is Sandwich ELISA
Introduction
A sandwich ELISA measures antigen between two layers of antibodies (capture and detection antibody). The target antigen must contain at least two antigenic sites capable of binding to antibodies.
Monoclonal or polyclonal antibodies can be used as the capture and detection antibodies in sandwich ELISA systems. Monoclonal antibodies recognize a single epitope that allows quantification of small differences in antigen.
A polyclonal is often used as the capture antibody to pull down as much of the antigen as possible. Sandwich ELISAs remove the sample purification step before analysis and enhance sensitivity (2–5 times more sensitive than direct or indirect).
Sandwich ELISA procedures can be difficult to optimize and tested match-paired antibodies should be used. This ensures the antibodies are detecting different epitopes on the target protein and do not interfere with the other antibody binding.
We are unable to guarantee our antibodies in sandwich ELISA unless they have been specifically tested.
Coating with capture antibody
(1)Coat the wells of a PVC microtiter plate with the 100ul of capture antibody at 1–10 μg/mL concentration in PBS.
(2) Unpurified antibodies (eg ascites fluid or antiserum or culture supernatant) may require increased concentration of the sample protein (try 10 μg/mL) to compensate for the lower concentration of specific antibody.
(3)Cover the plate with adhesive plastic and incubate overnight at 4°C.
(4)Remove the coating solution and wash the plate twice by filling the wells with 200 μL PBS containing 0.5% tween 20 (wash buffer).
Blocking and adding samples
(1)Block the remaining protein-binding sites in the coated wells by adding 200 μL blocking buffer (5% non-fat dry milk/PBS or 1% BSA in PBS) per well.
(2)Cover the plate with adhesive plastic and incubate for at least 1 h at 37°C.
(3)Wash the plate twice with 200 µL wash buffer.
(4)Add 100 μL of diluted samples to each well. Always compare signal of unknown samples against those of a standard curve. Run standards (duplicates or triplicates) and blank with each plate. Incubate for 1h at 37°C.
(5)Remove samples and wash the plate twice with 200 μL PBS.
Incubation with detection and secondary antibody
(1)Add 100 μL of diluted detection antibody to each well.
Check that the detection antibody recognizes a different epitope on the target protein to the capture antibody. This prevents interference with antibody binding. Use a tested matched pair whenever possible.
(2)Cover the plate with adhesive plastic and incubate for 1 h at 37°C.
(3)Wash the plate four times with wash buffer.
(4)Add 100 μL of conjugated secondary antibody diluted in PBS.
(5)Cover the plate with adhesive plastic and incubate for 1 h at 37°C.
(6)Wash the plate four times with PBS.
Detection
(1)Add 100 μL of the substrate solution (TMB) to each well.
(2)Wait for 5-10 min.
(3)After the colour development, stop the enzymatic reaction by adding 100 μL 1M sulfuric acid to the wells. Then, measure the absorbance in a microplate reader within 30 min of adding the stop solution






 Guidence of GeneMedi's protocol / procedure for the diagnostics application:
Guidence of GeneMedi's protocol / procedure for the diagnostics application: 