CRISPR/Cas9 Knockout Adenovirus Product: Adv-CMV-spCas9-EGFP Cat. No.: GMV-Crispr-AdV003
Order Information
| Product | Adv Vector Production Scale | Price(In USD) | Qty (Quantity) | Sum(In USD) |
|---|---|---|---|---|
| Adv-CMV-spCas9-EGFP | 1E+10PFU | 2820 | ||
| Adv-CMV-spCas9-EGFP | 1E+11PFU | 8100 | ||
| Adv-CMV-spCas9-EGFP | 1E+12PFU | 34500 | ||
| Shipping Cost: | 760.00 | |||
| Total: | ||||
Introduction to CRISPR/Cas9 Knockout Adenovirus Vectors
CRISPR/Cas9 premade products
| Cat No. | Products Name | Type of Crispr | Viral vector | Promoter | Fluorescent/Resistance | Tag | Order |
| GMV-Crispr-AAV001 | AAV-CMV-saCas9 | saCas9 | AAV | CMV | Null | HA |  |
| GMV-Crispr-AdV001 | Ad-CMV-spCas9 | spCas9 | Adenovirus | CMV | zsgreen | FLAG |  |
| GMV-Crispr-AdV002 | Adv-CMV-spCas9-zsgreen | spCas9 | Adenovirus | CMV | EGFP | FLAG |  |
| GMV-Crispr-AdV003 | Adv-CMV-spCas9-EGFP | spCas9 | Adenovirus | CMV | Null | FLAG |  |
| GMV-Crispr-AdV004 | Adv-CMV-spCas9-mcherry | spCas9 | Adenovirus | CMV | mcherry | FLAG |  |
| GMV-Crispr-LV001 | Lv-CMV-spCas9-puromycin | spCas9 | lentivirus | CMV | puromycin | FLAG |  |
| GMV-Crispr-LV002 | Lv-CMV-spCas9-zsgreen | spCas9 | lentivirus | CMV | zsgreen | FLAG |  |
| GMV-Crispr-LV003 | Lv-CMV-spCas9-EGFP | spCas9 | lentivirus | CMV | EGFP | FLAG |  |
| GMV-Crispr-LV004 | Lv-CMV-spCas9-mcherry | spCas9 | lentivirus | CMV | mcherry | FLAG |  |
| GMV-Crispr-LV005 | Lv-CBH-spCas9-puromycin | spCas9 | lentivirus | CBH | puromycin | FLAG |  |
| GMV-Crispr-LV006 | Lv-CBH-spCas9-zsgreen | spCas9 | lentivirus | CBH | zsgreen | FLAG |  |
| GMV-Crispr-LV007 | Lv-CBH-spCas9-EGFP | spCas9 | lentivirus | CBH | EGFP | FLAG |  |
| GMV-Crispr-LV008 | Lv-CBH-spCas9-mcherry | spCas9 | lentivirus | CBH | mcherry | FLAG |  |
Custom made CRISPR/Cas9 service
CRISPR/Cas9 User Manual
| Crispr/cas9 mediated Gene knockout  |
AAV Production CRISPR/Cas9 Knockout System-User Manual  |
Adenovirus CRISPR/Cas9 Knockout System-User Manual  |
Lentivirus-CRISPR/Cas9 Knockout System-User Manual  |
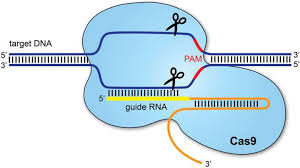
Recombinant adenovirus (Adv), a replication-defective adenoviral vector system, is widely used for gene delivery in most cell types. The adenoviral vectors provided by Genemedi are based on human adenovirus type 5 (Ad5), which is replication-incompetent (-E1/-E3) and can’t be integrated into host genome, guaranteeing the security for subsequent operations. Adv packaging by Genemedi shows almost 100% gene delivery in most cell types in the recombinant protein expression system both in vivo and in vitro. Derived from the type II CRISPR-Cas bacterial adaptive immune system, Cas9, an RNA-guided endonuclease, has emerged as a versatile genome-editing platform. Several microbial species possess this system, among which, Streptococcus pyogenes Cas9 (SpCas9), is the most robust and widely used Cas9 to date, primarily recognizes NGG PAMs and is consequently restricted to sites that contain this motif. By combining Adv packaging system with Cas9 genome-editing platform, Genemedi has further developed Adv-Cas9 system to achieve gene knockout with high efficiency, which is one of the top advanced gene knockout technology in the world.
Properties
| Adenovirus-Cas9 | |
|---|---|
| Quantity/Unit | Vials |
| Form | Frozen form |
| Sipping and Storage Guidelines | Shipped by dry ice, stored at -80 ° C, effective for 1 year. Avoid repeatedly freezing and thawing |
| Titer | > 1*10^10 PFU/ml |
Advantages
Applications and Figures
Quality control description
Technical Documents
 Adenovirus User Manuall on the Genemedi website.
2. For further information about how to design Adv-Cas9 system, please refer to the
Adenovirus User Manuall on the Genemedi website.
2. For further information about how to design Adv-Cas9 system, please refer to the  CRISPR/Cas9 Knockout Adenovirus User Manual on the Genemedi website.
CRISPR/Cas9 Knockout Adenovirus User Manual on the Genemedi website.
Frequently Asked Questions(FAQs)
- 1. Why does adenovirus have a relatively higher immunogenecity compared with rAAV?
-
AnswerThe adenovirus without E1/E3 can express all of the other genes in the viral backbone and hence induces immunogenic responses, while rAAV does not have any of the AAV genes, thus no immunogenicity from viral protein.
- 2. What is the role of the E1 Gene in adenoviruses?
-
AnswerSimply, the E1 gene products are early proteins that are transcribed in the early transcribed regions and required for proceeding subsequent steps in viral replication. The E1 gene contains E1A and E1B, involved in the replication of adenovirus. E1A is critical to start viral replication by promoting transcription from rep gene promoters, P5 and P19, and facilitate viral replication by activating the early adenovirus promoters.
- 3. How to store adenovirus?
-
AnswerIt would be better to store adenovirus in PBS at -80oC. Sucrose or DMSO may help to stabilize the vector.
- 4. How can you tell if your vector is lentiviral, retroviral, or adenoviral?
-
AnswerYou should blast your vector sequence and see if there’re sequences of reverse transcriptase and integrase (gene names: gag and pol), which are for lentiviral/retroviral vectors, but not for adenoviral. For another, if your plasmid is around 30-35kb in size, it's certainly adenoviral.
- 5. Is adenovirus a useful tool to study primary macrophage functions?
-
AnswerIn RAW264.7 and PM cells, adenovirus works very well, and it seems that IL1β expression is increased slightly after adenovirus transfection compared with negative control. While for the BMDM, adenovirus does not work, it may be better to use lentivirus instead, which gives a pretty good transduction efficacy and less inflammatory response.
- 6. Is it possible to infect a tissue preparation with lentivirus and afterwards with adenovirus and getting high efficiency in transduction?
-
AnswerIt is definitively possible to perform sequential transductions/infections. Polybreen, protamine sulfate or other transduction enhancing reagents are recommended to enhance viral particle infectivity.
- 7. Is it still possible to get the same mRNA accumulation in RT PCR after CRISPR/Cas9 induced gene deletion?
-
AnswerYes. The transcript will continue to be transcribed as nothing has changed with respect to the promoter of the gene. Besides the indel you introduced, the transcript will be unchanged and your primers (presumably not overlapping indel) will detect the transcript. You'll want to use Western blot to observe the effect of the indel.
- 8. I am trying to design gRNA for TLR4 gene CRISPR cas9 genome editing. TLR4 has multiple transcript variants. Which one should I choose?
-
AnswerYou should use a gRNA which target as many transcript variants as possible. To make it clear, target those exon(s) which can be found in the most variants of TLR4. As I see exon 1 and exon 4 are the best choices to target TLR4.
- 9. Which sequencing depth should I use for my CRISPR-Cas9 screen?
-
AnswerPCR amplification and Sanger sequencing are the main CRISPR KO screening methods.
- 10. Does the CRISPR/Cas9 system work for non-sequenced organisms?
-
AnswerIn theory it should be able to work with any organism. However, there may be some modifications that you will need to work out. You may need to codon optimize the Cas9 gene for your organism and confirm if there are many rare codons used in your organism. Next, you'll need to find a good promoter in your organism that you can use to make your crRNA. Maybe you'll need to add a nucleus localization signal to the Cas9 to make sure it stays in the nucleus, for Cas9 won't work if it is in the cytoplasm. Another issue you need to determine is what protospacer to use. If your organism is similar to one that is sequenced, then this may work, otherwise if you don't know the DNA sequence of your gene, it will be impossible to design a good protospacer against it. Lastly, you'll need a way to transfer Cas9 and crRNA into your organism, which should not hurt your organism.
- 11. Why is spCas9 the dominant CRISPR system?
-
AnswerOne of the main reasons is the relatively abundant PAM (protospacer adjacent motif) sequence of spCas9: NGG. This sequence is highly abundant throughout the genome, so this will give you a large amount of options for the sequence to target your sgRNA against. For instance, ST1 and NM require PAMs NNAGAAW and NNNNGATT respectively. These are far less abundant, and thus give far less targets in the genome to work with. SaCas9 is indeed a lot smaller than spCas9, and this can be Advantageous. However, its optimal PAM is NNGRRT. The second is that SpCas9 outperforms Sa/Nm in generating DSBs in the genome. The third part of the reason is also for historical reasons. There may be Cas9 species with other 3nt PAM sites. However, since the first one that was discovered was SpCas9, most systems have made adaptations of this Cas9 protein, so sgRNAs (and sgRNA design tools) can be used for multiple systems (including dCas9 activation / inhibition).
Reference
1. Hu J, L Zhang, Y Yang, Y Guo, Y Fan, M Zhang, W Man, E Gao, W Hu, RJ Reiter, H Wang and D Sun. (2017). Melatonin alleviates postinfarction cardiac remodeling and dysfunction by inhibiting Mst1. J Pineal Res 62.
2. Xu C, F Wu, C Mao, X Wang, T Zheng, L Bu, X Mou, Y Zhou, G Yuan, S Wang and Y Xiao. (2016). Excess iodine promotes apoptosis of thyroid follicular epithelial cells by inducing autophagy suppression and is associated with Hashimoto thyroiditis disease. J Autoimmun 75:50-57.
3. Wang Y, X Zhao, X Wu, Y Dai, P Chen and L Xie. (2016). microRNA-182 Mediates Sirt1-Induced Diabetic Corneal Nerve Regeneration. Diabetes 65:2020-31.
4. Luo T, J Fu, A Xu, B Su, Y Ren, N Li, J Zhu, X Zhao, R Dai, J Cao, B Wang, W Qin, J Jiang, J Li, M Wu, G Feng, Y Chen and H Wang. (2016). PSMD10/gankyrin induces autophagy to promote tumor progression through cytoplasmic interaction with ATG7 and nuclear transactivation of ATG7 expression. Autophagy 12:1355-71.
5. Liu TY, XQ Xiong, XS Ren, MX Zhao, CX Shi, JJ Wang, YB Zhou, F Zhang, Y Han, XY Gao, Q Chen, YH Li, YM Kang and GQ Zhu. (2016). FNDC5 Alleviates Hepatosteatosis by Restoring AMPK/mTOR-Mediated Autophagy, Fatty Acid Oxidation, and Lipogenesis in Mice. Diabetes 65:3262-3275.
6. Liu H, S Fang, W Wang, Y Cheng, Y Zhang, H Liao, H Yao and J Chao. (2016). Macrophage-derived MCPIP1 mediates silica-induced pulmonary fibrosis via autophagy. Part Fibre Toxicol 13:55.
7. Yao T, X Ying, Y Zhao, A Yuan, Q He, H Tong, S Ding, J Liu, X Peng, E Gao, J Pu and B He. (2015). Vitamin D receptor activation protects against myocardial reperfusion injury through inhibition of apoptosis and modulation of autophagy. Antioxid Redox Signal 22:633-50.
8. Wang B, A Ma, L Zhang, WL Jin, Y Qian, G Xu, B Qiu, Z Yang, Y Liu, Q Xia and Y Liu. (2015). POH1 deubiquitylates and stabilizes E2F1 to promote tumour formation. Nat Commun 6:8704.
9. Hua F, K Li, JJ Yu, XX Lv, J Yan, XW Zhang, W Sun, H Lin, S Shang, F Wang, B Cui, R Mu, B Huang, JD Jiang and ZW Hu. (2015). TRB3 links insulin/IGF to tumour promotion by interacting with p62 and impeding autophagic/proteasomal degradations. Nat Commun 6:7951.
10. Wang X, J Liu, J Zhen, C Zhang, Q Wan, G Liu, X Wei, Y Zhang, Z Wang, H Han, H Xu, C Bao, Z Song, X Zhang, N Li and F Yi. (2014). Histone deacetylase 4 selectively contributes to podocyte injury in diabetic nephropathy. Kidney Int 86:712-25.








